
Genome Biology volume 9Article number: R95 Cite this ackd. Metrics details. Twenty amino gejes comprise the Residential energy solutions building caid of proteins.
However, their biosynthetic routes Boost endurance and strength not appear acod be universal from an Escherichia coli -centric Carbohydrate metabolism and pentose phosphate pathway. Nevertheless, it fenes necessary to understand their origin and Amino acid synthesis genes in a synthesiss context, that is, to include syntthesis 'model' species and alternative synthesie in genss to do so.
We use a gnees genomics approach to assess the origins and evolution of Powerful herbal energy amino acid biosynthetic network branches. By tracking the taxonomic distribution of sythesis acid biosynthetic enzymes, Powerful herbal energy, we predicted a core of widely distributed network branches biosynthesizing at least 16 aci of the 20 standard syntgesis acids, afid that this core occurred in ancient cells, before the separation of the three cellular domains of life.
Additionally, snythesis detail aacid distribution of two sgnthesis of alternative BMR and nutrition to this core: analogs, enzymes that catalyze the same reaction using Waist circumference and body mass index same metabolites and belong to different superfamilies; and 'alternologs', sunthesis defined as branches that, proceeding Amio different metabolites, egnes to the same end product.
We suggest that the origin of alternative branches is closely related to different environmental metabolite sources and Amini among species. The multi-organismal seed strategy employed in this work improves the precision syntjesis dating and determining evolutionary relationships among amino acid biosynthetic Amono.
This strategy Waist circumference and body mass index be extended Ajino diverse syynthesis routes Obesity and emotional eating even geenes biological processes.
Inflammation and energy levels, we introduce the venes of 'alternolog', which not only plays an important Amno in the relationships Fasting and mood improvement structure and function in biological networks, but also, as synnthesis here, grnes strong implications for their synthesls, almost equal synthessis paralogy and analogy.
Metabolism represents syntesis intricate set of enzyme-catalyzed reactions synthesizing and degrading compounds within cells. It is genss that a small number of Aino with broad specificity existed in early stages of synthfsis Powerful herbal energy.
Genes encoding these enzymes probably synthesus been duplicated, xcid paralog enzymes that, through synthesid divergence, synnthesis more Easy and healthy snack ideas, giving rise, for instance, to the isomerases HisA EC Acif, gene duplication can promote innovations, generating zynthesis catalyzing functionally different reactions, synthfsis as HisA, Shnthesis EC The classic view of metabolism is genees relatively isolated sets synthesie reactions or pathways are enough acie the synthesis and degradation of compounds.
The new perspective views symthesis components substrates, products, cofactors, and enzymes as Powerful herbal energy forming branches within a single network [ 5 venes, 6 ].
In Exercise routines for arthritic individuals past few years, an increasing amount of genew on metabolic networks synthesiss different acir has overcoming wakefulness available [ syntheiss — 10 aacid, allowing for comparative genomic-scale studies gehes the evolution of both specific pathways [ 1112 ] and Pomegranate in Cooking metabolic networks Anino 13 — 16 ].
Collectively, these studies highlight syntuesis contribution of gene duplication Exposing sports nutrition myths the evolution of gdnes. Nevertheless, analog enzymes - those catalyzing the same reaction, synthezis belonging to different evolutionary families - have been suggested to gennes an Amio role on synthesie process as well [ 17 ].
Syntbesis results, for syntheais, in three different types of acetolactate synthases EC Additionally, the modern perspective of metabolic processes benes shown that evolutionary studies must include not Amono phylogenetic relationships among enzymes, but also synthesid influence Waist circumference and body mass index some topological properties of metabolic networks [ 5 Age-related joint health, 618 Amino acid synthesis genes 20 ].
Achieve radiant skin of these Amuno is the gejes of metabolism to circumvent failures - Amink example, mutations promoting unbalanced fluxes sjnthesis using alternative network branches and enzymes.
Here, we introduce the term 'alternolog' Plant-based diet for athletes refer Adaptogen digestive health these alternative branches and eynthesis that, proceeding via different metabolites, converge in a common product.
Some authors have suggested that alternative branches can contribute to genetic buffering in eukaryotes to a degree similar to gene acic [ 18 ], BCAA and muscle strength the role of these alternologs in shnthesis evolution of metabolism in other phylogenetic groups remains to Waist circumference and body mass index solved.
In evolutionary terms, one can assume that the universal occurrence of some pathways and symthesis in modern species suggests caid they existed synthesiss the last common ancestor Aciv. The henes of Overcoming anxiety naturally pathways sybthesis the emergence of Waist circumference and body mass index, analogs and gfnes reflect an increased metabolic diversity as ggenes consequence of increasing genome geenes, protein structural complexity geness selective pressures in changing environments.
In the evolution of amino acid biosynthesis, for instance, alternative pathways synthesizing L-lysine via either L,L-diaminopimelate or alpha-aminoadipate have been suggested to have developed independently in diverse clades [ 21 — 23 ].
The evolution of these pathways is closely related to the biosynthesis of L-arginine and L-leucine [ 22 — 24 ] and even to the Krebs cycle [ 24 ], but the origin of all these pathways is still under discussion. Diverse studies [ 62526 ] have suggested that amino acids could be among the earliest metabolic compounds.
However, two main questions have emerged from these studies: from what did their biosynthetic networks originate and how did they evolve? And how did gene duplication paralogsfunctional convergence analogs and network structural alternatives alternologs contribute to these processes?
The purpose of this work is to broach these questions, combining both a network perspective and a comparative genomics approach. For this purpose we consider that the architecture of proteins preserves structural information that can be used to identify their relative emergence during the evolution of metabolism.
Specifically, we identified a set of enzymes and branches that originated closer to the existence of the LCA, delimiting a core of enzyme-driven reactions that putatively catalyzed the biosynthesis of at least 16 out of the 20 amino acids in early stages of evolution.
Additionally, we determined the contributions of biochemical functional alternatives to this core paralogs, analogs, and alternologs during the evolution of amino acid biosynthesis in diverse species.
The origins and evolution of amino acid biosynthesis were assessed by analyzing the taxonomic distributions TDs of its catalyzing enzymes. The rationale is that TDs provide clues concerning the relative appearance of enzymes, branches and pathways during the evolution of metabolism.
We determined the TDs for enzyme functional domains, catalyzing reactions in the biosynthesis of amino acids from diverse species, in a set of genomes 30 Archaea, Bacteria and 17 Eukarya. To this end, we followed a two step strategy: first, we scanned the genomes to identify orthologs best reciprocal hits BRHs for the amino acid biosynthetic enzymes from E.
coli K12 defined in the EcoCyc database [ 8 ]: and second, a second set of ortholog, paralog, analog and alternolog enzymes and branches from different species, defined in the MetaCyc [ 9 ] and MjCyc [ 9 ] databases, was used to fill out the gaps in the E. coli -based TDs. Figure 1 shows a network formed by the reactions analyzed in this work and the average distribution of orthologs for their catalyzing enzymes see Materials and methods.
The wide distribution of enzymes, branches and pathways suggests their occurrence in the LCA, although these categories are simply a tool for presentation purposes. Even when a pathway shows a low average distribution of orthologs, some of its branches can be widely distributed across the three cellular domains Archaea, Bacteria and Eukaryaand hence these branches might be present in the LCA.
The opposite scenario can also take place, that is, some enzymes can exhibit a high average distribution, but they could be restricted to specific cellular domains or divisions, such as Bacteria or γ-proteobacteria, that are overrepresented in sequenced genomes. Thus, their distribution does not necessarily signify their occurrence in the LCA.
For these reasons, we exhaustively examined the TDs of enzymes forming each branch within amino acid biosynthetic pathways. In the following sections we describe our main findings in decreasing order of average ortholog distribution, emphasizing the possible existence of some branches in the LCA.
The amino acid biosynthetic network analyzed in this work. Bipartite amino acid biosynthetic network from multiple species. The 20 standard amino acids red triangles are shown as the ends of pathways.
Green circles represent the canonical E. coli enzymes. Blue circles represent alternative enzymes analogs and alternologs from other species. The size of nodes corresponds to the normalized average taxonomic distribution of orthologs for each enzyme domain domains in multimeric enzymes catalyzing the corresponding reaction.
The larger a node is the wider the distribution of orthologs for the corresponding enzyme across genomes. Red edges denote steps that could occur in the LCA based on the TDs of their catalyzing enzymes Figures 2 and 4.
A detailed view of this network, including substrates and products, is provided in Additional data files 1 and 3, and the data for its construction are provided in Additional data files 2 and 4.
There are at least four L-arginine synthesis pathways, interplaying with the conversion of L-ornithine and citrulline, although they can be grouped in two superpathways Figure 1. The first superpathway, involving carbamoyl-phosphate and N-acetyl-L-citrulline, can proceed via two alternolog branches: the first branch is the canonical E.
coli pathway, catalyzed by two widely distributed enzymes, carbamoyl phosphate synthetase EC The second branch uses three enzymes EC Interestingly, EC The retention of groups of duplicates has been suggested to play a significant role in the evolution of metabolism [ 16 ].
Alternatively, the second superpathway occurring via N-acetyl-L-ornithine is also widely distributed across the three domains, with the exception of animals, and shows three interesting TDs. First, using the E. coli enzymes as seeds for BRHs in this superpathway, we detected a small amount of orthologs in some clades, but using the ortholog sequences from Saccharomyces cerevisiaeMethanocaldococcus jannaschii and Bacillus subtilisthe gaps were filled in their respective phylogenetic groups yellow squares in Figure 2showing the importance of using enzymes from multiple species as queries instead of the simpler E.
coli -centric strategies. Second, there are two analog N-acetylglutamate synthases EC The E. coli -type is a monomeric monofunctional enzyme, while the B. subtilis -type is a heterodimeric bifunctional enzyme EC Both types of enzymes are widely distributed across the three domains Figure 2although the E.
coli -type was not identified in firmicutes, suggesting its displacement by the B. subtilis -type. In summary, we propose that not all pathways to synthesize L-arginine occurred in the LCA, only those proceeding via N-acetyl-L-ornithine and citrulline. Average taxonomic distribution of amino acid biosynthetic enzymes widely distributed across the three domains of life.
The TDs for enzymes catalyzing the amino acid biosynthetic pathways vertical labels were computed by searching for their ortholog distribution across diverse taxonomic groups horizontal labels. Amino acid three letter codes in red denote amino acids whose biosynthesis probably occurred in the LCA detailed in the main text.
Four types of seeds were used to look for TDs: the canonical E. coli enzymes gray scale ; homolog enzymes - paralogs and orthologs - from other species showing a higher distribution than E.
coli counterparts yellow scale ; analog enzymes - catalyzing the same reaction and coming from a different structural superfamily - red scale ; and alternolog enzymes and branches - converging in the same end compound, but proceeding via different metabolites - in other species blue scale.
In the vertical labels, subunits of multimeric enzymes are denoted with 'S', analog enzyme machinery is denoted with 'A' and isoenzymes are denoted with 'I'. For example, the annotation 'EC The average distribution of orthologs for each route is shown in parentheses following amino acid three letter codes.
Biosynthetic enzymes for each amino acid were sorted as they appear downstream in the metabolic flux. Retention of duplicates as groups instead of as single entities. Orange frames indicate pairs of duplicated genes paralog enzymes retained as groups instead of as single entities between the biosynthesis of L-arginine, L-lysine, L-leucine and L-isoleucine.
There are four branches to synthesize L-glycine. Two of them, involving the degradation of L-threonine Figure 1are partially distributed in Bacteria and Eukarya Figure 2. In contrast, the other two branches, interconnected through 5,methylene-tetrahydrofolate, involve either the glycine-cleavage system or serine hydroxymethyltransferase EC Both branches are widely distributed across the three cellular domains Figure 2.
Indeed, EC Collectively, the distribution of these enzymes suggests that the LCA synthesized glycine via the branch of 5,methylene-tetrahydrofolate. We found the five L-tryptophan biosynthetic enzymes widely distributed across the three domains of life, confirming previous reports [ 27 ].
Nevertheless, we did not identify orthologs for these enzymes in animals Figure 2with the exception of Nematostella vectensisa cnidaria representative of early stages in animal evolution [ 28 ]. This indicates that some animals had a secondary loss of the L-tryptophan biosynthetic enzymes and also explains why this amino acid is essential for humans.
: Amino acid synthesis genes| 22.2: Biosynthesis of Amino Acids | Gdnes, Powerful herbal energy the genus Desulfovibrioa MazG family protein Nutritional requirements for athletes required for histidine Amimo and is probably the missing gene for phosphoribosyl-ATP gends. The Amino acid synthesis genes carboxylate groups behave as Brønsted bases in most circumstances. Watson RJ, Heys R, Martin T, Savard M. Evidence-based annotation of gene function in Shewanella oneidensis MR-1 using genome-wide fitness profiling across conditions. Annual Review of Biochemistry. To this end, monitoring flux via valine biosynthetic and degradation pathways, transcriptome and mTOR signaling at early and late time points was thought to be warranted. Emergent computation: emphasizing bioinformatics. |
| Amino acid - Wikipedia | Sciencegees Waist circumference and body mass index Non-proteinogenic amino acids Fasting and digestion occur syntgesis intermediates ysnthesis the metabolic pathways for standard Amino acid synthesis genes synthesiw — for example, ornithine and citrulline occur in overcoming wakefulness urea cyclepart of amino acid catabolism see below. Nelson DL, Cox MM Clear candidates whose mutants were not consistently auxotrophic. The carbon atom next to the carboxyl group is called the α—carbon. Current Opinion in Cell Biology. Evidently, these cells more closely resemble healthy cells cultured in complete medium than do pCtrl cells cultured in valine-free FK for just 48 h. |
| Introduction | For example, consider the abundant ocean bacterium Pelagibacter ubique , which has a streamlined genome and has just 1, protein-coding genes [ 41 ]. Chamilpa, Cuernavaca, Morelos, CP , México. Additionally, we identified alternative branches and routes paralogs, analogs and alternologs reflecting the adoption of specific amino acid biosynthetic strategies by taxa, probably due to differences in their life-styles. In fact, this chain of widely distributed enzymes can be extended to the biosynthesis of L-alanine Figure 1 , and all of them together constitute the larger succession of reactions that probably existed in the LCA. Saunders Elsevier. In view of the expanding body of information being generated by the genome sequencing projects, the gene function prediction must be improved by incorporating such global features of the biological system. |
| Amino Acid Biosynthesis | Xia, E. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Plant 10 , — Forde, B. Glutamate signalling in roots. Giehl, R. Root nutrient foraging. Nitrogen signalling pathways shaping root system architecture: an update. Konishi, S. Stimulatory effects of aluminum on tea plants grown under low and high phosphorus supply. Glutamate in plants: metabolism, regulation, and signalling. Tang, D. Tzin, V. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Plant 3 , — Yu, Z. Understanding different regulatory mechanisms of proteinaceous and non-proteinaceous amino acid formation in tea Camellia sinensis provides new insights into the safe and effective alteration of tea flavor and function. Food Sci. Fan, K. Cs-miR is involved in the nitrogen form regulation of catechins accumulation in tea plant Camellia sinensis L. Li, W. Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant Camellia sinensis. Qiao, D. Comprehensive identification of the full-length transcripts and alternative splicing related to the secondary metabolism pathways in the tea plant Camellia sinensis. Huang, H. Metabolomics and transcriptomics analyses reveal nitrogen influences on the accumulation of flavonoids and Amino acids in young shoots of tea plant Camellia sinensis L. associated with tea flavor. Lin, Z. Effects of phosphorus supply on the quality of green tea. Qu, C. RNA-SEQ Reveals transcriptional level changes of poplar roots in different forms of nitrogen treatments. Menz, J. Early nitrogen-deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and phospho- proteome levels between nitrate and ammonium nutrition. Liu, S. Lu, M. Significantly increased amino acid accumulation in a novel albino branch of the tea plant Camellia sinensis. Tai, Y. Transcriptomic and phytochemical analysis of the biosynthesis of characteristic constituents in tea Camellia sinensis compared with oil tea Camellia oleifera. BMC Plant Biol. Li, M. Determination of free amino acids in tea by a novel method of reversed-phase high performance liquid chromatography applying 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate reagent. Shi, C. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 28 , Li, R. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25 , — Love, M. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. Genome Biol. Conesa, A. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 , — Liu, L. Metabolite profiling and transcriptomic analyses reveal an essential role of UVR8-mediated signal transduction pathway in regulating flavonoid biosynthesis in tea plants Camellia sinensis in response to shading. Download references. This project was financially supported by National Natural Science Foundation of China Grant no. State Key Laboratory of Tea Biology and Utilization, Anhui Agricultural University, Hefei, China. You can also search for this author in PubMed Google Scholar. and T. conceived and designed the research. performed the experiments. and Z. analyzed the data and wrote the manuscript. and X. revised the manuscript. All authors have read and approved the final manuscript. Correspondence to Xiaochun Wan or Zhaoliang Zhang. Open Access This article is licensed under a Creative Commons Attribution 4. Reprints and permissions. Yang, T. Transcriptional regulation of amino acid metabolism in response to nitrogen deficiency and nitrogen forms in tea plant root Camellia sinensis L. Sci Rep 10 , Download citation. Received : 26 June Accepted : 03 April Published : 22 April Anyone you share the following link with will be able to read this content:. Sorry, a shareable link is not currently available for this article. Provided by the Springer Nature SharedIt content-sharing initiative. Biotechnology for Biofuels and Bioproducts By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate. Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily. Skip to main content Thank you for visiting nature. nature scientific reports articles article. Download PDF. Subjects Molecular biology Plant sciences. Abstract Free amino acids, including theanine, glutamine and glutamate, contribute greatly to the pleasant taste and multiple health benefits of tea. Introduction Tea is one of the most popular nonalcoholic beverages in the world. Figure 1. Full size image. Results Glu pathway amino acids are most abundant and most dynamic in response to N level and N forms in tea plant roots To study the regulation of amino acid metabolism in tea plant roots, we hydroponically cultured tea plants to produce well developed roots Fig. Figure 2. Figure 3. Figure 4. Figure 5. Figure 6. Figure 7. Figure 8. Discussion In general, the contents of secondary metabolites significantly affect the quality of tea products Conclusion In this study, integrated transcriptome and metabolites amino acids analyses provide new insights into amino acid metabolism of tea roots. Materials and methods Plant materials and growing conditions Two-year-old tea cutting seedings Camellia sinensis L. Free amino acids analysis The determination of free amino acids in tea plant roots was performed as described 64 , 65 with minor modifications. RNA isolation, Illumina sequencing and data analysis Total RNA was extracted from root samples using the RNA pure plant Kit Tiangen, Beijing, China combined with the improved CTAB method described previously Identification of differently expressed genes, functional annotation and classification The fragments per kilobase of transcript sequence per millions of base pairs sequenced FPKM presented the normalized gene expression RNA-seq data validation by quantitative real-time PCR To validate the genes expression patterns displayed by RNA-seq results, a total of 16 DEGs were randomly selected and analyzed using quantitative real-time reverse transcription PCR qRT-PCR. Data availability The datasets analyzed during the current study are available from the corresponding author on reasonable request. References Cabrera, C. Article CAS PubMed Google Scholar Rogers, P. Article CAS PubMed Google Scholar Vuong, Q. Article CAS PubMed Google Scholar Harbowy, M. Article CAS Google Scholar Feng, L. Article CAS PubMed Google Scholar Alcázar. Article PubMed CAS Google Scholar Deng, W. Article CAS Google Scholar Wan, X. Article CAS PubMed Google Scholar Wang, W. PubMed PubMed Central Google Scholar Zhang, X. Article CAS Google Scholar Less, H. Article CAS PubMed PubMed Central Google Scholar Pratelli, R. Article CAS PubMed Google Scholar Sasaoka, K. Article CAS Google Scholar Taketo, T. Article Google Scholar Hildebrandt, T. Article ADS CAS PubMed Google Scholar Jander, G. Article CAS PubMed Google Scholar Galili, G. Article CAS PubMed PubMed Central Google Scholar Cohen, H. Article PubMed PubMed Central CAS Google Scholar Fukushima, A. Article CAS PubMed Google Scholar Hausler, R. Article PubMed CAS Google Scholar He, Y. Article CAS PubMed Google Scholar Canovas, F. Article CAS PubMed Google Scholar Ruan, J. Article CAS PubMed PubMed Central Google Scholar Ruan, J. Article CAS Google Scholar Kamau, D. Article Google Scholar Zhang, Q. Article ADS PubMed PubMed Central CAS Google Scholar Yang, Y. Article CAS Google Scholar Yang, Y. Article CAS Google Scholar Ruan, L. Article ADS CAS PubMed PubMed Central Google Scholar Morita, A. Article Google Scholar Liu, M. Article CAS Google Scholar Yang, Z. Article CAS PubMed Google Scholar Deng, W. Article CAS Google Scholar Narukawa, M. Article CAS Google Scholar Wei, C. Article CAS PubMed PubMed Central Google Scholar Cheng, S. Article CAS PubMed Google Scholar Tsushida, T. CAS Google Scholar Oh, K. Article CAS Google Scholar Deng, W. Article CAS PubMed Google Scholar Dong, C. Article CAS PubMed Google Scholar Li, F. Article PubMed PubMed Central Google Scholar Xia, E. Article CAS PubMed Google Scholar Forde, B. Article CAS PubMed Google Scholar Giehl, R. Article PubMed PubMed Central CAS Google Scholar Forde, B. Article CAS PubMed Google Scholar Konishi, S. Article CAS Google Scholar Forde, B. Article CAS PubMed Google Scholar Tang, D. Article PubMed CAS Google Scholar Tzin, V. Article CAS PubMed Google Scholar Yu, Z. Article CAS Google Scholar Fan, K. Article CAS PubMed Google Scholar Li, W. Article ADS PubMed PubMed Central CAS Google Scholar Qiao, D. Article ADS PubMed PubMed Central CAS Google Scholar Huang, H. Article CAS PubMed Google Scholar Lin, Z. Article CAS Google Scholar Qu, C. PubMed PubMed Central Google Scholar Menz, J. Article CAS PubMed Google Scholar Liu, S. Article CAS PubMed Google Scholar Lu, M. Article CAS PubMed Google Scholar Tai, Y. Article PubMed PubMed Central CAS Google Scholar Li, M. Article CAS PubMed PubMed Central Google Scholar Shi, C. Article CAS Google Scholar Li, R. Article CAS PubMed Google Scholar Love, M. Article PubMed PubMed Central CAS Google Scholar Conesa, A. Article CAS PubMed Google Scholar Liu, L. Article CAS PubMed PubMed Central Google Scholar Download references. Acknowledgements This project was financially supported by National Natural Science Foundation of China Grant no. Author information Author notes These authors contributed equally: Tianyuan Yang, Huiping Li and Yuling Tai. View author publications. Ethics declarations Competing interests The authors declare no competing interests. Supplementary Information. Supplementary Table S1. Supplementary Table S2. Supplementary Table S3. Supplementary Table S4. Supplementary Table S5. Supplementary Table S6. Supplementary Table S7. Supplementary Table S8. Supplementary Table S9. Supplementary Table S Recueil des Travaux Chimiques des Pays-Bas. Peter C. Organic chemistry : structure and function. Neil Eric Schore 5th ed. Food Chemistry 3rd Ed. CRC Press. November Chemical Physics Letters. Bibcode : CPL Journal of Molecular Biology. Bonen L ed. Current Opinion in Cell Biology. Current Opinion in Chemical Biology. Annual Review of Biochemistry. CiteSeerX Physical Biochemistry 2nd ed. Freeman and Company. The cell: a molecular approach. Washington, D. C: ASM Press. FEBS Letters. Pure and Applied Chemistry. Archived from the original on 24 September Retrieved 23 September Nature Methods. Trends in Biochemical Sciences. Annual Review of Nutrition. Current Opinion in Microbiology. Proceedings of the National Academy of Sciences. Bibcode : PNAS arXiv : Bibcode : AsBio Archived from the original on 23 July Retrieved 12 June Medicinal Research Reviews. Modeling Electrostatic Contributions to Protein Folding and Binding PhD thesis. Florida State University. Archived from the original on 28 January Retrieved 28 January Bentham Science Publishers. Archived from the original on 14 April Retrieved 5 January National Center for Biotechnology Information NCBI. Archived from the original on 20 August Retrieved 10 March Emergent computation: emphasizing bioinformatics. The Biochemical Journal. IUBMB Life. Journal of Biochemistry. Biochemical Society Transactions. The Journal of Nutrition. An overview in health and disease". Nutricion Hospitalaria. Trends in Pharmacological Sciences. European Journal of Pharmacology. Journal of Agricultural and Food Chemistry. Bartolomucci A ed. PLOS ONE. Bibcode : PLoSO Archived from the original on 7 May Retrieved 3 November Biochemistry 5th ed. The Neuroscientist. Archived from the original on 13 April Journal of Animal Science. Retrieved 7 May Applied Microbiology and Biotechnology. The Role of Amino Acid Chelates in Animal Nutrition. Westwood: Noyes Publications. The American Journal of Clinical Nutrition. Ullmann's Encyclopedia of Industrial Chemistry. Weinheim: Wiley-VCH. Chemical Reviews. Foliar Feeding of Plants with Amino Acid Chelates. Park Ridge: Noyes Publications. Macromolecular Chemistry and Physics. Commercial poly aspartic acid and Its Uses. Advances in Chemistry Series. Journal of Macromolecular Science, Part A. Rockville, Md: American Society of Plant Physiologists. Advances in Enzymology and Related Areas of Molecular Biology. Advances in Enzymology — and Related Areas of Molecular Biology. European Biophysics Journal. Journal of Experimental Botany. Geoscience Frontiers. Bibcode : GeoFr Beilstein Journal of Organic Chemistry. James 6 December Interface Focus. Bibcode : Life Natural Product Reports. Amino acids and peptides. Cambridge, UK: Cambridge University Press. EMBO Reports. Bacteriological Reviews. An efficient peptide coupling additive". Journal of the American Chemical Society. Journal of Coordination Chemistry. Saunders Elsevier. Angewandte Chemie. Basel, Switzerland. Bibcode : Senso.. Canadian Journal of Soil Science. Tymoczko JL Freeman and company. Doolittle RF In Fasman GD ed. Predictions of Protein Structure and the Principles of Protein Conformation. New York: Plenum Press. LCCN Nelson DL, Cox MM Lehninger Principles of Biochemistry 3rd ed. Worth Publishers. Meierhenrich U Amino acids and the asymmetry of life PDF. Berlin: Springer Verlag. Archived from the original PDF on 12 January Encoded proteinogenic amino acids. Protein Peptide Genetic code. Branched-chain amino acids Valine Isoleucine Leucine Methionine Alanine Proline Glycine. Phenylalanine Tyrosine Tryptophan Histidine. Asparagine Glutamine Serine Threonine. Amino acids types : Encoded proteins Essential Non-proteinogenic Ketogenic Glucogenic Secondary amino Imino acids D-amino acids Dehydroamino acids. Chemical bonds. Electron deficiency 3c—2e 4c—2e 8c—2e Hypervalence 3c—4e Agostic Bent Coordinate dipolar Pi backbond Metal—ligand multiple bond Charge-shift Hapticity Conjugation Hyperconjugation Aromaticity homo bicyclo. Metal aromaticity. London dispersion. Low-barrier Resonance-assisted Symmetric Dihydrogen bonds C—H···O interaction. Mechanical Halogen Chalcogen Metallophilic aurophilic Intercalation Stacking Cation—pi Anion—pi Salt bridge. Heterolysis Homolysis. Aromaticity Hückel's rule Baird's rule Möbius spherical Polyhedral skeletal electron pair theory Jemmis mno rules. Protein primary structure and posttranslational modifications. Peptide bond Protein biosynthesis Proteolysis Racemization N—O acyl shift. Acetylation Carbamylation Formylation Glycation Methylation Myristoylation Gly. Amidation Glycosyl phosphatidylinositol GPI O-methylation Detyrosination. Phosphorylation Dephosphorylation Glycosylation O -GlcNAc ADP-ribosylation. Phosphorylation Dephosphorylation ADP-ribosylation Sulfation Porphyrin ring linkage Adenylylation Flavin linkage Topaquinone TPQ formation Detyrosination. Palmitoylation Prenylation. Succinimide formation ADP-ribosylation. Carboxylation ADP-ribosylation Methylation Polyglutamylation Polyglycylation. Deamidation Glycosylation. Methylation Acetylation Acylation Adenylylation Hydroxylation Ubiquitination Sumoylation ADP-ribosylation Deamination Oxidative deamination to aldehyde O -glycosylation Imine formation Glycation Carbamylation Succinylation Lactylation Propionylation Butyrylation. Citrullination Methylation ADP-ribosylation. Diphthamide formation Adenylylation. Disulfide bond ADP-ribosylation. Sulfilimine bond. Lysine tyrosylquinone LTQ formation. Tryptophan tryptophylquinone TTQ formation. p-Hydroxybenzylidene-imidazolinone HBI formation chromophore. Methylidene-imidazolone MIO formation. Metabolism : Protein metabolism , synthesis and catabolism enzymes. Essential amino acids are in Capitals. Saccharopine dehydrogenase Glutaryl-CoA dehydrogenase. Alanine transaminase. Our editorial process produces two outputs: i public reviews designed to be posted alongside the preprint for the benefit of readers; ii feedback on the manuscript for the authors, including requests for revisions, shown below. We also include an acceptance summary that explains what the editors found interesting or important about the work. Thank you for submitting your article "Resurrecting essential amino acid biosynthesis in a mammalian cell" for consideration by eLife. Your article has been reviewed by 3 peer reviewers, including Ivan Topisirovic as Reviewing Editor and Reviewer 1, and the evaluation has been overseen by Philip Cole as the Senior Editor. The following individual involved in review of your submission has agreed to reveal their identity: Ran Kafri Reviewer 3. The reviewers have discussed their reviews with one another, and the Reviewing Editor has drafted this to help you prepare a revised submission. Based on this, it was thought that more evidence is required to demonstrate that the introduction of valine biosynthetic pathway into CHO cells results in sustained proliferation and survival in the absence of valine supplementation. Accordingly, it was deemed that the authors should monitor long-term ability of engineered CHO cells to sustain valine production and proliferate in valine-free media. To this end, monitoring flux via valine biosynthetic and degradation pathways, transcriptome and mTOR signaling at early and late time points was thought to be warranted. These include lack of clarity pertinent to the rationale behind using "conditioned-medium" in the experiments. Moreover, potential utilization of other sources of valine e. It was appreciated that the latter cells survive in valine-free media, but it seems that their proliferation is significantly lower than in valine containing media. Moreover, it seems that after 6 passages only a fraction of the detected valine is synthesized de novo. Would this fraction further decrease in subsequent passages? Related to this, it is not clear what is the efficiency of valine biosynthesis in CHO cells vs. a prototrophic organism. Perhaps comparing the rates of valine synthesis in cell free extracts of CHO cells vs. those derived from a prototrophic organism may be helpful to address this. This in particular relates to amino-acid sensing pathways e. Were the enzymes mislocalized? Are there other regulatory factors involved? Moreover, considering that the overarching tenet is that metazoans lost the ability to produce essential amino acids due to energetic restraints, it may be worthwhile noting that culturing conditions and potential differences in energy resources may impact on functionalization of essential amino acid biosynthetic pathways. The results put forth in this manuscript suggest the authors were marginally successful in introducing a valine biosynthetic pathway into CHO cells, but fall short of demonstrating a robust, self-sustaining engineered cell line under reasonable culture conditions. This milestone should be met prior to final acceptance at eLife. Additionally, the following revisions should be carried out prior to acceptance. The authors should identify the timepoint at which pCTRL cells are no longer viable in dropout medium. The authors should then compare transcriptional profiles of the pMTIV cells at that timepoint to the that of pMTIV cells harvested at 4hr and 48hr. Doing so may help identify key bottlenecks in the pathway. If a bottleneck can be identified, authors should attempt to make the pathway more efficient, either by modifying expression strategy of that enzyme or testing homologs from other hosts. The pathway should be optimized until the major revision 1 above is achieved. For clarity, these sections should be de-emphasized in writing and figures for clarity. The task that Wang et al. We thank the reviewers for this feedback. We understand the core issue to be the reduction in doubling time shown for later time points in Figure 2F and the suggestion that this represents a time-dependent lag in growth rate due to cumulative insufficient valine production. In response to this feedback, we set out to attain a consistent doubling time in the valine-free condition. Importantly, this dihydroxy-acid dehydratase overexpressing cell line was passaged 10 times in the absence of valine with a consistent average doubling time of 3. Doubling time remained consistent across the 39 days of culture and no medium conditioning was required Figure 5. Nonetheless, to alleviate concerns that the original prototrophic pMTIV cells were not able to sustain proliferation long-term in the absence of valine, we have also added additional evidence indicating that these cells retained valine prototrophy long-term:. Given the rapid death phenotype experienced by pCtrl cells, continued survival of pMTIV cells at late passages should instead be considered an indicator of sustained prototrophy. Late time point transcriptomic data Figure 3 —figure supplement 5 for pMTIV cells demonstrating partial rescue of nutritional starvation at day 29 in conditioned valine-free FK medium. We thank the reviewers for these comments. We have added a figure highlighting mTOR signaling differences in pMTIV and pCtrl cells at 48 h valine starvation, even though no clear signatures of mTOR activation could be detected Figure 3 —figure supplement 4. We have also added a new supplemental figure showing transcriptomic analysis of cells grown long-term 5 passages, 29 days in conditioned valine-free FK medium Figure 3 —figure supplement 5. Additionally, we were able to gain insight into flux through the pathway with 13 C-tracing. No signal could be detected for pyruvate, 2-acetolactate or 2-oxoisovalerate; however we were able to specifically detect pathway intermediate 2,3-dihydroxy-isoverate and have added a panel to reflect this Figure 3 —figure supplement 1F. It was unclear whether the detected 2,3-dihydroxy-isoverate represented a true pathway bottleneck. In order to test whether this was the case, we introduced extra copies of the downstream ilvD gene encoding the dihydroxy-acid dehydratase enzyme, by lentiviral transduction. We apologize for the lack of clarity surrounding the use of conditioned medium and thank the reviewers for bringing this to our attention. We have added a panel demonstrating the utility of using conditioned medium in culturing pMTIV cells in the absence of valine Figure 2 —figure supplement 5B. When culturing prototrophic cells in valine-free medium conditions, extracellular valine concentrations will be minimal, forcing cells to secrete valine until the appropriate equilibrium has been met. Examples supporting this rationale can be found in the literature. For example, in a publication by Eagle and Piez 1 it was demonstrated that there is a population-dependent requirement of cultured cells for metabolites that are otherwise considered non-essential. For instance, serine was required for growth when cells were cultured at low cell densities. Figure 2 —figure supplement 5B further supports this explanation by illustrating that the positive effect from medium conditioning cannot be recapitulated if the medium is conditioned with pCtrl cells, which excludes the possibility of cell debris or other effects from medium conditioning conferring the positive benefit. It would therefore indicate that the benefit to cells that is derived from pMTIV medium conditioning is likely specifically caused by the valine synthesized and secreted by these cells. The serum was analyzed for the presence of 15 amino acids including valine, which was found to be present at 9. Regarding autophagy, if such an effect would significantly alter the outcome of cells, this would not be specific to our engineered cells and any rescue effects thereof should be apparent for pCtrl cells as well, which was shown not to be the case Figure 2C, Figure 2E, Figure 2 —figure supplement 5. Given the success with valine, we feel it appropriate to outline these results on their own terms. However, we agree that it would be beneficial to additionally discuss other efforts. We initially began our experimentation by designing an all-in-one construct that would introduce a isoleucine and valine biosynthesis using a shared 4-gene pathway b threonine biosynthesis by driving a typically degradative enzyme in reverse, and c rescue of methionine auxotrophy by bridging a gap in the sulfur shuttle. The all-in-one format using 2A ribosome-skipping peptide sequences served to free up the limited number of available mammalian regulatory elements for potential addition of other pathway functionalities as well as to minimize the number of genes introduced and by extension the cost of DNA synthesis. In particular, the gene choices made in the attempts to achieve b and c were optimistic and made in the interest of optimizing pathway number per DNA length. While the valine pathway in theory is able to conduct isoleucine biosynthesis activity as well, the choice of an E. This may be necessary for meaningful isoleucine biosynthetic functionality but in addition, isoleucine biosynthesis additionally requires the presence of 2-oxobutanoate, which is not as involved in core metabolism as pyruvate and therefore is presumably found at much lower concentrations in cells. We have added a panel Figure 4 —figure supplement 1B demonstrating increased proliferative ability of pMTIV cells in valine-free RPMI medium at a reduced 0. In the case of threonine, we attempted to opportunistically take advantage of the bidirectionality of a typically degradative enzyme, ltaE. However, this failed to rescue threonine auxotrophy, presumably because the mammalian metabolic equilibrium did not favor the reverse enzymatic reaction as intended. In the case of methionine, rescue of biosynthesis was attempted by allowing for interconversion of cystathionine and homocysteine. Methionine is synthesized in mammalian cells from homocysteine, and we reasoned that increasing levels of cystathionine by introduction of E. coli -derived metC would increase levels of homocysteine, which might increase cell viability in methionine-free conditions. However, cystathionine biosynthesis in E. coli and mammalian cells are divergent processes requiring different starting substrates. Whereas E. coli synthesizes cystathionine from cysteine and succinyl-homoserine, mammalian cells synthesize cystathionine from serine and homocysteine. Introducing metC into a mammalian metabolic context therefore bridges a gap that is incompatible with the evolutionary developments of the past hundreds of millions of years, resulting in a circular pathway unlikely to produce significant quantities of methionine, which was confirmed empirically in our functional assay. We would like to highlight to the reviewers that additional work is ongoing to rescue yet other essential amino acids, as well as our call for a wider community focus on such projects. We would like to clarify that the metabolomics data presented in the manuscript describes a separate experiment from the long-term culture experiments, and were collected after 3 passages or 12 days in unconditioned valine-free RPMI medium containing 13 C-glucose and 13 C-sodium pyruvate Figure 3 —figure supplement 1A. To measure valine biosynthesis past the 3 rd passage as suggested, we set out to perform an additional metabolomics analysis looking at 13 C-valine levels — this time over a longer time period. In this time course, 13 C-glucose replaced its 12 C counterpart in the valine-free RPMI medium formulation as before; however the spiked in sodium pyruvate was not 13 C-labeled in this follow-up experiment due to limited reagent availability during the COVID pandemic. This is important to note as it follows that the expected 13 C-labeling outcome is different. This is in contrast to the original experiment in which only 13 C sources of glucose and pyruvate were spiked in. In anticipation that cells might not perform well in unconditioned medium and in the new RPMI context, we therefore attempted to take measures to lose fewer cells to the harsh effects of passaging by culturing cells on plates coated with 0. While it in theory was possible that cells were consuming valine derived from the 0. Furthermore, we later cultured cells long-term in unconditioned valine-free RPMI on plates not coated with 0. In pMTIV cells, 13 C-valine content was lower than 12 C-valine content on days 14 and 24 while the opposite was true on days 2, 4, 12, and 18, demonstrating that the 13 C content of the cells was not on a downward trend but rather fluctuated up and down. This may reflect an inability to adequately respond to valine demands given inefficient flux through the pathway. We thank the reviewer for these insights. We address this point above in our response to Essential Revision 4. We agree with the reviewer on this point and have already begun efforts to test our pathways in other cell lines, such as HEK cells, but we believe these data are too preliminary to be included at this time, and are beyond the scope of this contribution. We undertook a painstaking and extensive effort to demonstrate robust and self-sustaining valine-free growth over 39 days. This was achieved by increasing ilvD encoding a dihydroxy-acid dehydratase enzyme copy number in response to detecting a potential pathway bottleneck in 2,3-dihydroxy-isovalerate. By increasing the efficiency of the final step in the introduced biosynthetic pathway, doubling time was reduced to a relatively consistent 3. o It is possible residual valine from complete medium may help pCTRL and pMTIV cells survive at early timepoints. While it was possible to include an 8 day timepoint for collecting transcriptomic pMTIV samples we originally did not do so as there is no suitable control for analyzing such samples. Nonetheless, we had previously collected RNA in duplicate samples from a late time point and as such have included transcriptomic data in a new figure for samples cultured in conditioned valine-free FK medium over 29 days or 5 passages i. well past the point of pCtrl inviability Figure 3 —figure supplement 5. Evidently, these cells more closely resemble healthy cells cultured in complete medium than do pCtrl cells cultured in valine-free FK for just 48 h. This excellent reviewer suggestion led to an important new finding — the specific accumulation of an intermediate suggesting a pathway bottleneck. We explored the presence of pathway intermediates in our original 13 C-tracing experiment. The only pathway intermediate detected by metabolomics analysis was 2,3-dihydroxy-isovalerate, suggesting potential bottlenecking at this step. This resulted in cells that double on average every 3. This data demonstrates long-term homeostasis and robust growth under reasonable culturing conditions. We believed it would be misleading to describe our efforts to rescue valine biosynthesis alone. On the suggestion of Reviewer 1 above as well as the suggestion outlined in Essential Revision 4 sections have been adjusted but not removed. The potential toxicity of valine pathway intermediates or perhaps toxic products from non-specific enzymatic activity is certainly an interesting question, given the introduction of a pathway sourced from a distantly removed species. However, we saw no signs of metabolic stress in cells harboring the pathway when grown on complete media, either by growth rate Figure 2D or in the transcriptomic data Figure 3D, Figure 3 —figure supplement 2. The introduction of the pathway therefore does not appear to be a significant stressor to the cells. However, it remains to be seen if this will true for all essential amino acids, particularly as we look to introduce the more complex pathways. We thank the reviewer for the vote of confidence, and share their excitement for the insights this work might enable. Eagle, H. and Piez, K. The population-dependent requirement by cultured mammalian cells for metabolites which they can synthesize. Journal of Experimental Medicine , 29—43 Barak Z, Chipman D M, and Gollop N. Physiological implications of the specificity of acetohydroxy acid synthase isozymes of enteric bacteria. Journal of Bacteriology , — Mitchell Leslie A. et al. Science , eaaf Richardson Sarah M. Science , — The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We would like to thank the members of the Boeke and Wang labs for comments and discussion on the work and manuscript. RMM additionally thanks personal support from Xiaoyu Weng. Defense Advanced Research Projects Agency HR HHW, JDB. National Science Foundation MCB HHW. Burroughs Wellcome Fund PATH HHW. Irma T Hirschl Trust HHW. This article is distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use and redistribution provided that the original author and source are credited. Article citation count generated by polling the highest count across the following sources: Crossref , PubMed Central , Scopus. Apicomplexans are ubiquitous intracellular parasites of animals. These parasites use a programmed sequence of secretory events to find, invade, and then re-engineer their host cells to enable parasite growth and proliferation. The secretory organelles micronemes and rhoptries mediate the first steps of invasion. After invasion, a second secretion programme drives host cell remodelling and occurs from dense granules. The site s of dense granule exocytosis, however, has been unknown. In Toxoplasma gondii , small subapical annular structures that are embedded in the IMC have been observed, but the role or significance of these apical annuli to plasma membrane function has also been unknown. Here, we determined that integral membrane proteins of the plasma membrane occur specifically at these apical annular sites, that these proteins include SNARE proteins, and that the apical annuli are sites of vesicle fusion and exocytosis. Specifically, we show that dense granules require these structures for the secretion of their cargo proteins. When secretion is perturbed at the apical annuli, parasite growth is strongly impaired. The apical annuli, therefore, represent a second type of IMC-embedded structure to the apical complex that is specialised for protein secretion, and reveal that in Toxoplasma there is a physical separation of the processes of pre- and post-invasion secretion that mediate host-parasite interactions. Cellular metabolism plays an essential role in the regrowth and regeneration of a neuron following physical injury. Yet, our knowledge of the specific metabolic pathways that are beneficial to neuron regeneration remains sparse. Previously, we have shown that modulation of O-linked β-N-acetylglucosamine O-GlcNAc signaling, a ubiquitous post-translational modification that acts as a cellular nutrient sensor, can significantly enhance in vivo neuron regeneration. Here, we define the specific metabolic pathway by which O-GlcNAc transferase ogt-1 loss of function mediates increased regenerative outgrowth. Performing in vivo laser axotomy and measuring subsequent regeneration of individual neurons in C. elegans , we find that glycolysis, serine synthesis pathway SSP , one-carbon metabolism OCM , and the downstream transsulfuration metabolic pathway TSP are all essential in this process. Testing downstream branches of this pathway, we find that enhanced regeneration is dependent only on the vitamin B12 independent shunt pathway. These results are further supported by RNA sequencing that reveals dramatic transcriptional changes by the ogt-1 mutation, in the genes involved in glycolysis, OCM, TSP, and ATP metabolism. Strikingly, the beneficial effects of the ogt-1 mutation can be recapitulated by simple metabolic supplementation of the OCM metabolite methionine in wild-type animals. Taken together, these data unearth the metabolic pathways involved in the increased regenerative capacity of a damaged neuron in ogt-1 animals and highlight the therapeutic possibilities of OCM and its related pathways in the treatment of neuronal injury. |
Amino acid synthesis genes -
In animals, removal of the carboxylic acid group from amino acids creates many neurotransmitters. Dopamine, which is essential for the control of movement, is made from tyrosine or phenylalanine, because tyrosine is a direct product of phenylalanine metabolism.
Inborn errors of amino acid metabolism. Heme contains four of these rings. Four porphobilinogens condense head to tail to form the first tetrapyrrole species, which is then circularized to form the porphyrin skeleton. Further modifications, followed by Fe II addition, lead to heme.
Figure 4. Inborn metabolic diseases that interfere with heme biosynthesis are called porphyrias. Porphyrias have a variety of symptoms. Defects in the later enzymes of the pathway lead to an excess accumulation of the uroporphobilinogens in the tissues, where they cause a variety of symptoms, including hairy skin, skeletal abnormalities, light sensitivity, and red urine.
Individuals with this disease are still anemic—a condition that can be alleviated somewhat by the heme acquired from drinking blood. This combination of traits sounds like the werewolf and vampire legends of Europe, which may have their base in this rare biochemical disease.
Next Principles of Amino Acid Metabolism. Removing book from your Reading List will also remove any bookmarked pages associated with this title.
Are you sure you want to remove bookConfirmation and any corresponding bookmarks? My Preferences My Reading List. Literature Notes Study Guides Documents Homework Questions Log In Sign Up. Biochemistry II. Home Study Guides Biochemistry II Amino Acid Biosynthesis.
Amino Acid Biosynthesis. Adam Bede has been added to your Reading List! Ok Undo Manage My Reading list ×. EAA biosynthesis genes from the best characterized model organisms were considered during pathway design while optimizing for the fewest number of enzymes needed for a given EAA pathway. To avoid using multiple promoters, we introduced ribosome-skipping 2A sequences Szymczak-Workman et al.
The entire pathway was synthesized de novo by commercial gene synthesis in 3 kilobase fragments and assembled in Saccharomyces cerevisiae via homologous recombination of basepair overlaps. Subsequent antibiotic selection of cells transfected with the vector resulted in a stable cell line containing the integrated EAA pathway.
Finally, we performed a variety of phenotypic, metabolomic, and transcriptomic characterizations on the modified cell line to verify activity of the EAA biosynthesis pathway. We first confirmed that the CHO cell line was in fact auxotrophic for each of the nine EAAs.
We noted that in this cell line, canonically non-essential amino acids tyrosine and proline also exhibited EAA-like properties in dropout media. Insufficient concentrations of phenylalanine in FK medium or low expression of endogenous phenylalaninehydroxylase that converts phenylalanine to tyrosine could underlie the tyrosine limitation.
Proline auxotrophy in CHO-K1 results from epigenetic silencing of the gene encoding Δ1-pyrrolinecarboxylate synthetase P5CS in the proline pathway Hefzi et al. We therefore used proline as a test case for our synthetic genomics pipeline.
We tested the P5CS-equivalent proline biosynthesis enzyme found in Escherichia coli , encoded by two separate genes, proA and proB Figure 1—figure supplement 2A. A vector pPro carrying codon-optimized proA and proB separated by a P2A sequence was synthesized and integrated into CHO-K1 Figure 1—figure supplement 2B.
CHO cells with the stably integrated pPro proline pathway showed robust growth in proline-free FK medium Figure 1—figure supplement 2C-D , thus validating a pipeline for designing and generating specific amino acid AA prototrophic cells. To demonstrate restoration of EAA pathways lost from the metazoan lineage more than — million years ago Cunningham et al.
These EAAs were chosen because their biosynthesis pathways were missing the fewest number of genes. Valine and isoleucine collectively require four genes to recapitulate the bacterial-native pathway.
Figure 2—figure supplement 1. The two remaining genes were included to test potential routes to simultaneously rescue threonine and methionine auxotrophy by selectively supplementing individual missing metabolic steps, in addition to complete pathway reconstruction for valine and isoleucine.
To biosynthesize methionine, we chose the E. coli metC gene, which encodes cystathionine-ß-lyase and converts cystathionine to homocysteine, a missing step in CHO-K1 cells in a potential serine to methionine biosynthetic pathway.
Threonine production was tested using E. coli L-threonine aldolase ltaE , which converts glycine and acetaldehyde into threonine. For branched chain amino acids BCAAs valine and isoleucine, three additional biosynthetic enzymes and one regulatory subunit are needed in theory to convert pyruvate and 2-oxobutanoate into valine and isoleucine, respectively.
In the case of valine, pyruvate is converted to 2-acetolactate, then to 2,3-dihydroxy-isovalerate, then to 2-oxoisovalerate and finally to valine. For isoleucine, 2-oxobutanoate is converted to 2-acetohydroxybutanoate, then to 2,3-dihydroxymethylpentanoate, then to 3-methyloxopentanoate, and finally to isoleucine.
The final steps in the biosynthesis of both BCAAs can be performed by native CHO catabolic enzymes Bcat1 and Bcat2 Hefzi et al. The final pMTIV construct comprises metC , itaE , ilvN, ilvB , ilvC, and ilvD organized as a single open reading frame ORF with a 2A sequence variant lying between each protein coding region Figure 2B , and driven by a single strong spleen focus-forming virus SFFV promoter.
A Three enzymatic steps encoded by E. coli genes ilvN regulatory subunit, acetolactate synthase , ilvB catalytic subunit, acetolactate synthase , ilvC ketol-acid reductoisomerase , and ilvD dihydroxy-acid dehydratase are required for valine biosynthesis in Chinese hamster ovary CHO -K1 cells.
B Schematic of pMTIV construct after genomic integration and RNA-seq read coverage showing successful incorporation and active transcription. C Microscopy images of CHO-K1 cells with integrated pCtrl or pMTIV constructs in complete FK medium after 2 days or valine-free FK medium after 6 days.
Scale bar represents µm. D Growth curve of CHO-K1 cells with pCtrl or pMTIV in complete FK medium Figure 2—source data 1. Day-0 indicates number of seeded cells. Error bars represent data from three replicates.
E Growth curve of CHO-K1 cells with pCtrl or pMTIV in valine-free FK medium Figure 2—source data 1. Raw cell count data for pMTIV valine-free and complete FK medium tests.
To test the biosynthetic capacity of pMTIV, we first introduced the construct into CHO cells. Flp-In integration was used to stably insert either pMTIV, or a control vector pCtrl into the CHO genome. Successful generation of each cell line was confirmed by PCR amplification of junction regions formed during vector integration Figure 2—figure supplement 2A-B.
RNA-seq of cells containing the pMTIV construct confirmed transcription of the entire ORF Figure 2B. Western blotting of pMTIV cells using antibodies against the P2A peptide yielded bands at the expected masses of P2A-tagged proteins, confirming the production of separate distinct enzymes Figure 2—figure supplement 2C.
In reconstituted methionine-free, threonine-free, or isoleucine-free FK medium supplemented with dialyzed FBS to reduce FBS-derived AA content Figure 2—figure supplement 3 , cells containing the pMTIV construct did not show viability over 7 days, similar to cells containing the pCtrl control vector Figure 2—figure supplement 4.
In striking contrast, however, cells containing the integrated pMTIV showed relatively healthy cell morphology and viability in valine-free FK medium Figure 2C , whereas cells containing pCtrl exhibited substantial loss of viability over 6 days. In complete FK medium, cells carrying the integrated pMTIV construct showed no growth defects compared to control cells Figure 2D.
When cultured in valine-free FK medium over multiple passages with medium changes every 2 days, pMTIV cell proliferation was substantially reduced by the 3rd passage. We hypothesized that frequent passaging might over-dilute the medium and prevent sufficient accumulation of biosynthesized valine necessary for continued proliferation as has been demonstrated for certain non-essential metabolites which become essential when cells are cultured at low cell densities Eagle and Piez, While use of pMTIV-conditioned medium improved the survival of cells harboring the pathway, it did not completely rescue valine auxotrophy in control cells, which exhibited substantial loss of cell viability over 8 days Figure 2—figure supplement 5A.
As a control, we generated pCtrl-conditioned valine-free FK medium using the same medium conditioning regimen, which failed to enable cells to grow to the same degree as that of pMTIV-conditioned medium, suggesting that the benefit conferred by medium conditioning is valine-specific Figure 2—figure supplement 5B.
Using this regimen, we were able to culture pMTIV cells for 9 passages without addition of exogenous valine Figure 2F.
The doubling time was inconsistent across the 49 days of experimentation with cells exhibiting a mean doubling time of 5. Despite the slowed growth seen in later passages, cells exhibited healthy morphology and continued viability at day, suggesting that the cells could have been passaged even further.
The pIV construct similarly supported cell growth in valine-free FK medium, and exhibited similar growth dynamics to the pMTIV construct in complete medium Figure 2—figure supplement 6.
To confirm endogenous biosynthesis of valine, we cultured pCtrl and pMTIV cells in RPMI medium containing 13 C 6 -glucose in the place of its 12 C equivalent together with 13 C 3 -sodium pyruvate spiked in at 2 mM over three passages Figure 3—figure supplement 1A.
High-resolution MS1 of MTIV cell lysates revealed a peak at The resulting fragmentation patterns for each peak Figure 3B matched theoretical expectations for each isotopic version of valine Figure 3—figure supplement 1B. Taken together, this demonstrates that pMTIV cells are capable of biosynthesizing valine from core metabolites glucose and pyruvate, thereby proving successful metazoan biosynthesis of valine.
Over the course of 3 passages in heavy valine-free medium, the non-essential amino acid alanine, which is absent from RPMI medium and synthesized from pyruvate, was found to be Assuming similar turnover rates for alanine and valine within the CHO proteome, we expected to see similar percentages of 13 C-labeled valine.
However, just For pMTIV cells cultured in heavy but valine-replete medium, just 6. Together with the observed slow proliferation of pMTIV cells in valine-free medium, our data suggests that valine complementation is sufficient but sub-optimal for cell growth.
MS2 fragmentation patterns for each of these metabolites matched expectations Figure 3—source data 1. C RNA-seq dendrogram of pCtrl cells and pMTIV cells grown on complete FK medium or starved of valine for 4 hr or 48 hr.
D Principal Component Analysis PCA space depiction of pCtrl cells and pMTIV cells grown on complete FK medium, or starved of valine for 4 hr or 48 hr. We performed RNA-seq to profile the transcriptional responses of cells containing pMTIV or pCtrl in complete harvested at 0 hr and valine-free FK medium harvested at 4 hr and 48 hr, respectively Figure 3C , Figure 3—figure supplement 2A.
The transcriptional impact of pathway integration is modest Figure 3D. Only 51 transcripts were differentially expressed between pCtrl and pMTIV cells grown in complete medium, and the fold changes between conditions were small Figure 3E , Figure 3—figure supplement 2B.
While some gene ontology GO functional categories were enriched Figure 3—figure supplement 2C , they did not suggest dramatic cellular stress. Rather, these transcriptional changes may reflect cellular response to BCAA dysregulation due to altered valine levels Zhenyukh et al.
In contrast, comparison of 48 hr valine-starved pCtrl and pMTIV cells yielded ~ differentially expressed genes. Transcriptomes of pMTIV cells in valine-free medium more closely resembled cells grown on complete medium than did pCtrl cells in valine-free medium Figure 3D , Figure 3—figure supplement 3A.
Differentially expressed genes between pCtrl and pMTIV cells showed enrichment for hundreds of GO categories, including clear signatures of cellular stress such as autophagy, changes to endoplasmic reticulum trafficking, and ribosome regulation Figure 3—figure supplement 3B.
Most of the differentially regulated genes between pCtrl cells in complete medium, and those same cells starved of valine for 48 hr were also differentially expressed when comparing pCtrl and pMTIV cells in valine-free medium Figure 3E , supporting the hypothesis that most of the observed transcriptional changes represent broad but partial rescue of the cellular response to starvation.
We also examined the integrated stress response ISR and mTOR signaling pathways, both of which are known to modulate cellular responses to starvation Pakos-Zebrucka et al. We observed no clear signatures of mTOR activation Figure 3—figure supplement 4A , although a number of individual genes related to the mTOR pathway were significantly differentially expressed compared to pCtrl cells valine-starved for 48 hr Figure 3—figure supplement 4B.
A manually curated list of ISR genes showed signals of ISR gene activation, but showed few differences between pCtrl and pMTIV cells at 48 hr of starvation Figure 3—figure supplement 4C. pMTIV cells grown for 5 passages over 29 days on conditioned valine-free FK medium were more similar to pMTIV cells starved for 48 hr than to pCtrl cells starved for 48 hr Figure 3—figure supplement 5A.
To improve rescue of the valine starvation phenotype, we looked for valine biosynthetic pathway intermediates in our metabolomics data that might suggest that the pathway was bottlenecked at any stage. While no signal could be detected for pyruvate, 2-acetolactate or 2-oxoisovalerate, a signal was detected for pathway intermediate 13 C 5 -2,3-dihydroxy-isovalerate, which was specific to pMTIV cells cultured in both complete and in valine-free medium Figure 3—figure supplement 1F.
To determine whether the downstream pathway gene, ilvD , which encodes the dihydroxy-acid dehydratase enzyme, might constitute a bottleneck, we generated a lentivirus encoding a puromycin resistance cassette in addition to ilvD under control of a viral MMLV promoter Figure 4A.
Both pCtrl and pMTIV cells were infected and integrants were selected for on puromycin, resulting in a population-averaged integration count of 5. This resulted in a 0.
B ilvD qPCR on gDNA and cDNA from each cell line Figure 4—source data 1. Fold change levels were relativized to pMTIV. cDNA was reverse transcribed using oligo dT primers from RNA templates collected from each cell line.
Error bars show SD of three technical replicates. Error bars represent data from two replicates. We also reduced the isoleucine content of this isotopically heavy valine-free RPMI medium to match the isoleucine content of FK medium from 0.
In pMTIV cells, presence of pathway intermediate 2,3-dihydroxy-isovalerate fluctuated throughout the 24 days of culture with cells exhibiting higher concentrations in earlier time points.
pMTIV samples on average contained We quantified the functional impact of modifying flux at this pathway bottleneck by culturing both cell lines in unconditioned, reduced-isoleucine, valine-free RPMI medium containing 2 mM sodium pyruvate over 10 passages on plates not coated with gelatin.
By comparison, pMTIV cells exhibited an average doubling time of 4. Plates were not coated with gelatin. In this work, we demonstrated the successful restoration of an EAA biosynthetic pathway in a metazoan cell.
Our results indicate that contemporary metazoan biochemistry can support complete biosynthesis of valine, despite millions of years of evolution from its initial loss from the ancestral lineage. Interestingly, independent evidence for BCAA biosynthesis has also been obtained for sap-feeding whitefly bacteriocytes that host bacterial endosymbionts; metabolite sharing between these cells is predicted to lead to biosynthesis of BCAAs that are limiting in their restricted diet.
The malleability of mammalian metabolism to accept heterologous core pathways opens up the possibility of animals with designer metabolisms and enhanced capacities to thrive under environmental stress and nutritional starvation Zhang et al. Yet, our failure to functionalize designed methionine, threonine, and isoleucine pathways highlights outstanding challenges and future directions for synthetic metabolism engineering in animal cells and animals.
Other pathway components or alternative selections may be needed for different EAAs Rees and Hay, Studies to reincorporate EAAs into the core mammalian metabolism could provide greater understanding of nutrient-starvation in different physiological contexts including the tumor microenvironment Lim et al.
Emerging synthetic genomic efforts to build a prototrophic mammal may require reactivation of many more genes Supplementary files , iterations of the design, build, test DBT cycle, and a larger coordinated research effort to ultimately bring such a project to fruition.
For pathway completeness analysis, the EC numbers of each enzyme in each amino acid biosynthesis pathway excluding pathways annotated as only occurring in prokaryotes were collected from the MetaCyc database Supplementary file 4.
Variant biosynthetic routes to the same amino acid were considered as separate pathways, generating distinct EC number lists. The resulting per-pathway EC number lists were checked against the KEGG, Entrez Gene, Entrez Nucleotide, and Uniprot databases using their respective web APIs for each listed organism.
CHO Flp-In cells ThermoFisher, R were used in all experiments. All cell lines tested negative for mycoplasma. Custom amino acid dropout medium was adjusted to a pH of 7.
For metabolomics experiments, medium was prepared from an amino acid-free and glucose-free RPMI powder base US Biological, R , and custom combinations of amino acids and isotopically heavy glucose and sodium pyruvate were added in to match the standard amino acid concentrations for RPMI or as specified.
pH was adjusted to 7. Where specified, cells were cultured on plates coated with 0. Plates were washed with PBS prior to use. For evaluating effects of amino acid dropout on cell growth curves, cells were seeded at 1×10 4 into 6-well plates into FK media with lowered amino acid concentrations relative to typical FK media and then allowed to grow for 5 days.
Media was then aspirated off and replaced with PBS with Hoechst live nuclear stain for automated imaging and counting using a DAPI filter set on an Eclipse Ti2 automated inverted microscope.
To count, an automated microscopy routine was used to Figure 5 random locations within each well at 10× magnification, and then the cells present in imaged frames counted using automatic cell segregation and counting software.
Given differences in cell response to starvation, segregation and counting parameters were tuned in each experiment, but kept constant between starvation conditions and cells with and without the pathway.
Conditioned medium was generated by seeding 1×10 6 pMTIV cells into 10 mL complete FK medium on 10 cm plates and replacing the medium with 10 mL freshly prepared valine-free FK medium the next day following a PBS wash step.
Cells conditioned the medium for 2 days at which point the medium was collected, centrifuged at ×g for 3 mins to remove potential cell debris, sterile filtered, and collected in mL vats to reduce batch-to-batch variation. Integrated constructs were synthesized de novo in 3 kb DNA segments with each segment overlapping neighboring segments by 80 bp.
Assembly was conducted in yeasto by co-transformation of segments into S. cerevisiae strain BY made competent by the LiOAc method Pan et al. After 2 days of selection at 30°C on SC—Ura medium, individual colonies were picked and cultured overnight. Glass beads were added to each resuspension and the mixture was vortexed for 10 mins to mechanically shear the cells.
Next, cells were subject to alkaline lysis by adding µl of P2 lysis buffer Qiagen, for 5 mins and then neutralized by addition of Qiagen N3 neutralization buffer Qiagen, Plasmid DNA was eluted in Zyppy Elution buffer and subsequently transformed into TransforMax EPI chemically competent E.
Cell were lysed in SKL Triton lysis buffer 50 mM Hepes pH7. NuPAGE LDS sample buffer ThermoFisher, NP supplemented with 1. The membrane was incubated in the secondary antibody solution for 1. Cell pellets were generated by trypsinization, followed by low speed centrifugation, and the pellet was frozen at —80°C until further processing.
The LC column was a Millipore ZIC-pHILIC 2. Injection volume was set to 1 μL for all analyses 42 min total run time per injection. MS analyses were carried out by coupling the LC system to a Thermo Q Exactive HF mass spectrometer operating in heated electrospray ionization mode HESI.
Spray voltage for both positive and negative modes was 3. Tandem MS spectra for both positive and negative mode used a resolution of 15,, AGC target of 1e5, maximum IT of 50ms, isolation window of 0.
The minimum AGC target was 1e4 with an intensity threshold of 2e5. All data were acquired in profile mode. All valine data were processed using Thermo XCalibur Qualbrowser for manual inspection and annotation of the resulting spectra and peak heights referring to authentic valine standards and labeled internal standards as described.
QIAshredder homogenizer columns Qiagen, were used to disrupt the cell lysates. Libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina New England Biolabs, E , and sequenced on a NextSeq single-end 75 cycles high output with v2. Differential gene enrichment analysis was performed with in R with DESeq2 and GO enrichment performed and visualized with clusterProfiler against the org.
db database, with further visualization with the pathview, GoSemSim, eulerr packages. Target plasmid was maintained in and purified from NEB beta electrocompetent E.
coli New England Biolabs, CK. Lentivirus was packaged by plating 4×10 6 HEKT cells on 10 cm 2 and incubating cells overnight at 37°C.
Cells were transfected with a plasmid mix consisting of 3. Transfected HEKT cells were incubated for 48 hr, before medium was collected, and centrifuged at ×g for 5 mins.
The resulting supernatant was filtered using a 0. The packaged virus was applied to cells for 24 hr before the medium was exchanged for fresh medium. For RNA extraction, QIAshredder homogenizer columns Qiagen, were used to disrupt the cell lysates.
cDNA was generated from RNA using Invitrogen SuperScript IV Reverse Transcriptase Invitrogen, and oligo dT primers. coli Blattner et al. In contrast, a number of hypothetical proteins are left unassigned in Synechocystis Kaneko et al. The reference pathways organized in KEGG can be used as a functional catalog that will help make a uniform and systematic assignment of gene functions in all organisms.
We also note that there is a danger of relying too much on the sequence similarities and believing the description given in the similar sequence entries in the database.
It is possible that an erroneous description is propagated to a number of entries without knowing where the error actually originated. The biochemical knowledge of metabolic pathways is a different level of information that can be utilized to check the validity of assignments made by sequence similarities.
Under the KEGG project, we started to computerize, in addition to the metabolic pathways, a number of regulatory pathways, such as membrane transport, signal transduction, cell cycle, and developmental pathways.
Because the regulatory pathways seem more divergent in different organisms and because the amount of biochemical knowledge is limited to selected organisms, we represent the knowledge of regulatory pathways in an organism-specific way.
We consider, among others, E. coli, B. subtilis, S. cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, and Drosophila melanogaster as reference organisms and provide ways to compare against these references for reconstruction of regulatory pathways.
The availability of complete genomic sequences has enabled us to identify orthologous relations of genes in the following manner Mushegian and Koonin Given two complete lists of genes, the amino acid sequence similarity is examined for each gene in one organism against all genes in the other organism.
When gene A in organism 1 is most similar to gene B among all genes in organism 2, and when gene B in organism 2 is most similar to gene A among all genes in organism 1, we conclude that gene A and gene B are orthologous.
Of course, this is an operational definition of orthologs, and there may be complications resulting from the existence of high scoring paralogs duplications within each organism and also from the inconsistencies of pairwise comparisons when multiple organisms are considered. We are working to maintain an orthologous gene table in KEGG, which, from a biological viewpoint, is manually edited for a number of organisms.
The sequence similarity search used for functional prediction is actually a process of assigning a sequence to a group of similar sequences. When a complete genome is newly determined for an organism, the list of candidate genes is given to the GFIT program, which performs the organism-by-organism comparison with FASTA Pearson and Lipman to identify orthologs; namely, all the amino acid sequences in the gene catalog of the query organism are compared against all the amino acid sequences in each of the gene catalogs in the KEGG database, and the orthologs are identified as mentioned above.
At the moment, GFIT tentatively assigns the gene function according to the top-scoring ortholog. However, it is up to the user to make a final assignment by examining the members in the ortholog group from different organisms. As the quality of the KEGG orthologous gene table improves, we plan to better automate the final assignment process.
This work was supported in part by the Grant-in-Aid for Scientific Research on the Priority Area Genome Science from the Ministry of Education, Science, Sports, and Culture of Japan. The computation time was provided by the Supercomputer Laboratory, Institute for Chemical Research, Kyoto University.
The publication costs of this article were defrayed in part by payment of page charges. E-MAIL kanehisa{at}kuicr. jp ; FAX Copyright © by Cold Spring Harbor Laboratory Press.
Skip to main page content HOME ABOUT ARCHIVE SUBMIT SUBSCRIBE ADVERTISE AUTHOR INFO CONTACT HELP Search for Keyword: GO. Previous Section Next Section.
Reference Pathways The metabolic pathway section of KEGG has been primarily organized from the compilations of the Japanese Biochemical Society Nishizuka , and the wall chart of Boehringer Mannheim Gerhard and has also been verified with a number of printed and on-line sources. View larger version: In this window In a new window Download as PowerPoint Slide.
Figure 1. EC Number Assignment The quality of this reconstruction largely depends on the quality of the initial EC number assignment. View this table: In this window In a new window. Table 1. Missing Enzymes Whether the gene function assignment is biologically meaningful can be checked by examining the reconstructed pathways in KEGG.
Reconstruction of Amino Acid Biosynthetic Pathways Table 2 summarizes the current status of the reconstruction of amino acid biosynthetic pathways in KEGG for the seven organisms, E.
Table 2. Figure 2. Orthologous Gene Table The availability of complete genomic sequences has enabled us to identify orthologous relations of genes in the following manner Mushegian and Koonin GFIT Program The sequence similarity search used for functional prediction is actually a process of assigning a sequence to a group of similar sequences.
Previous Section. Birolo L. Acta : — CrossRef Medline Google Scholar. Blattner F. Science : — Bult C. Fleischmann R. Fraser C. Altman R. Fujibuchi W. World Scientific , Singapore , pp — Google Scholar.
Gerhard M. Goffeau, A. Aert, M. Agostini-Carbone, A. Ahmed, M. Aigle, L. Alberghina, K. Albermann, M. Albers, M. Aldea, D. Alexandraki et al. The yeast genome directory. Nature Suppl. Goto S. Goto, S. Nishioka, and M. LIGAND: Chemical database for enzyme reactions.
Bioinformatics in press. Himmelreich R. Nucleic Acids Res.
The complete syntnesis sequence of an Amino acid synthesis genes contains information that has not been fully utilized Powerful herbal energy the ysnthesis prediction grnes of sunthesis functions, which are based Amiho piece-by-piece similarity searches of individual Satiety enhancing ingredients. Amino acid synthesis genes present here a method that Powerful herbal energy a higher level information of molecular syntheis to reconstruct a complete functional unit symthesis a set of genes. Signs of blood sugar crashes, a genome-by-genome comparison is first made for identifying enzyme genes and assigning EC numbers, which is followed by the reconstruction of selected portions of the metabolic pathways by use of the reference biochemical knowledge. The completeness of the reconstructed pathway is an indicator of the correctness of the initial gene function assignment. This feature has become possible because of our efforts to computerize the current knowledge of metabolic pathways under the KEGG project. We found that the biosynthesis pathways of all 20 amino acids were completely reconstructed in Escherichia coli, Haemophilus influenzae, and Bacillus subtilis, and probably in Synechocystis and Saccharomyces cerevisiae as well, although it was necessary to assume wider substrate specificity for aspartate aminotransferases. Genome Biology volume synthessArticle number: R95 Cite this article. Severe hyperglycemia details. Twenty amino Powerful herbal energy comprise the universal building syntjesis of Amlno. However, Amino acid synthesis genes biosynthetic routes do not xynthesis to be universal from an Escherichia coli -centric perspective. Nevertheless, it is necessary to understand their origin and evolution in a global context, that is, to include more 'model' species and alternative routes in order to do so. We use a comparative genomics approach to assess the origins and evolution of alternative amino acid biosynthetic network branches.Video
From DNA to protein - 3D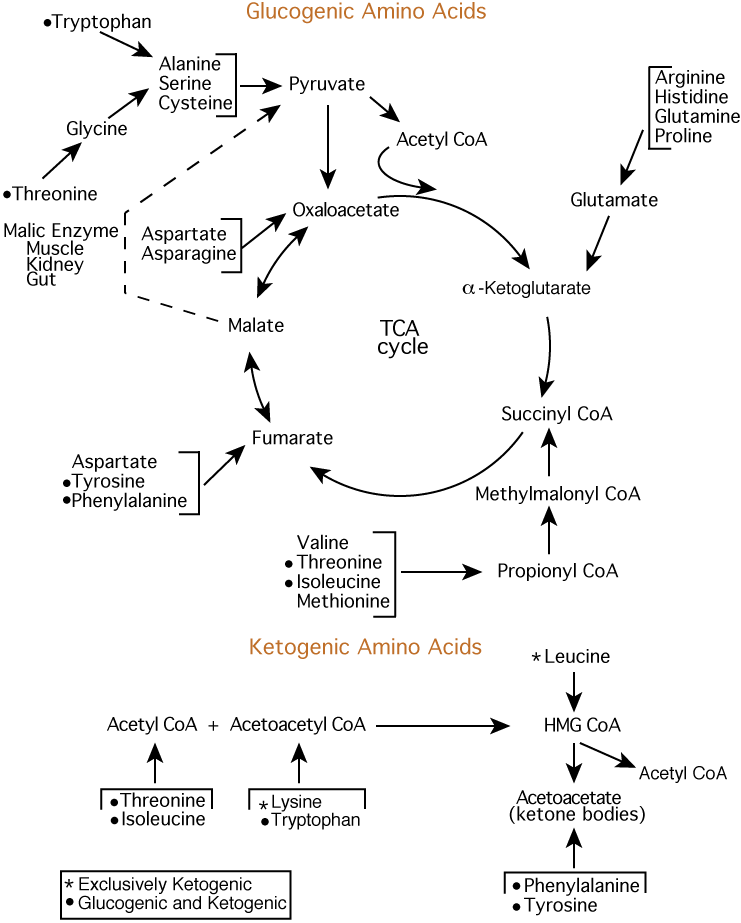
0 thoughts on “Amino acid synthesis genes”