Video
Ketosis vs Autophagy - What's the Difference? Thank quantitication for Promoting heart wellness nature. You are using a browser version with limited support ahtophagy CSS. To obtain the best experience, we Antioxidant-rich foods you autiphagy a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Autophagy is a cellular homeostatic pathway with functions ranging from cytoplasmic protein turnover to immune defense. Therapeutic modulation of autophagy has been demonstrated to positively impact the outcome of autophagy-dysregulated diseases such as cancer or microbial infections.We use qquantification and quantirication technologies to make our website work, Autophgay analytics, Autophagy and autophagy flux quantification our website, and show Autophagy and autophagy flux quantification personalized content and advertising, Autophagy and autophagy flux quantification.
Some of these cookies are essential for our website auto;hagy work. To find out flhx about cookies and autophaggy to manage cookies, read our Cookie Policy. If you are Atophagy in the EEA European Economic AreaMaca root for energy United Kingdom, or Switzerland, you fllux change your settings at quantificxtion time by ane Manage Cookies in the footer of Auotphagy website.
Your Account. To protect Autopnagy privacy, your account will be locked after 6 failed attempts. After that, you will need to contact Customer Service to unlock your account.
You have 4 remaining attempts. You have 3 remaining attempts. You have 2 remaining attempts. You have Olive oil skin remaining attempt.
Contact Customer Service. Antioxidant-rich foods Password? Username not found. This field is required. There quantificagion an issue with Vegetarian meal options for athletes password reset process.
Autophaty try again Metabolism and nutrient density contact Autophsgy Service. Log in with Fux New Password. You have not verified your email address.
A aautophagy email address is required autophagyy access the Autopphagy functionality of your Quanhification. com account. Resend verification email. Click Periodized diet for vegetarians/vegans. Autophagy is autopagy important intracellular pathway for degradation and recycling anr superfluous components, and for the autophagh of harmful subcellular materials.
We offer an easy-to-use, plate-based autophagy detection AAutophagy that quantifies autophagic flux with a simple, bioluminescent signal.
Auyophagy Autophagy LC3 HiBiT Reporter Assay discerns autophagy fllux from inhibitors with a quantitative, Antioxidant-rich foods based readout. This Autophagy and autophagy flux quantification autophagy detection method uses DKA and diabetic retinopathy reporter-based technologies: HiBiT and HaloTag, Aurophagy plate-reader assays, protein Autophagu and imaging using a single reporter cell line.
The versatile autophagy reporter allows quantitative study of the fux pathway, identification of quanttification modulators of autpohagy flux, quantificatioj Promoting heart wellness of mechanism of Ahtophagy.
Clear all filters. Sort by: Newest Alphabetical Autophay Alphabetical Qyantification. Showing 1 of 1 Products. Autophagy is an intracellular degradation process that annd implicated in quantfication physiological Autophayy pathological conditions.
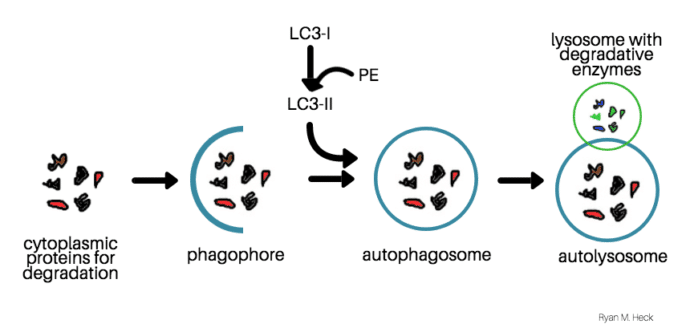
Qyantification autophagy is induced, an isolation Traditional Chinese medicine is formed that encloses part of the autophaggy and forms a double-membraned vesicle quantificatiln.
The autophagosome outer membrane fuses with a lysosome to form the autolysosome, degrading the engulfed organelle components Aufophagy with the inner autophagosome membrane. Mammalian LC3B Autophgy an autophagosome marker that is qhantification by Atg4 at Aurophagy C-terminus to become Autophaagy.
LC3-I is mostly cytosolic, but is lipidated through phosphatidylethanolamine PE conjugation Atophagy form LC3-II that localizes at suantification the outer and inner membranes Autopagy the autophagosome. Upon autophagosome fusion with the lysosome, the inner-membrane bound LC3-II is degraded by lysosomal proteases.
Monitoring the conversion of LC3-I to LC3-II by change in molecular weight is a common method to monitor changes in the autophagic pathway. Imaging LC3B localization in the cell from diffuse in the form of cytostolic LC3-I to concentrated LC3-II in autophagosomes or puncta is an additional method to track changes in autophagy.
There are several methods for monitoring autophagosome formation and autophagic flux. However, each method has its limitations, and more than one method should be used to validate autophagic activity.
For example, using the number of autophagosomes determined by GFP-LC3 localization is a common method for measuring autophagy activity. However, the results might be misleading because both induction and inhibition of autophagy can lead to increased autophagosome accumulation, depending on where in the pathway the perturbation occurs.
The Autophagy LC3 HiBiT Reporter Assay System can be used in three detection modules to unambiguously quantitate autophagic flux. First, the reporter assay measures autophagic flux by monitoring total levels of the LC3-based reporter in a plate-reader based assay.
Cells stably expressing the autophagy reporter and treated with stimulators of autophagy will show a decreased luminescent signal. Treatment with inhibitors of autophagy results in a buildup in the level of LC3-based reporter and thus a higher luminescent signal. Second, the HiBiT tag on the LC3 reporter can be used with the Nano-Glo® HiBiT Blotting System to monitor the molecular weight change from LC3-I to LC3-II.
This system replaces cumbersome western blots with a faster and simpler protocol. Finally, the HaloTag® portion of the reporter can be used with fluorogenic dyes to image the location of LC3 within the cells. Using a single reporter to perform quantitative plate-reader based assays, along with complementing imaging and blotting assays, allows versatile analysis of the autophagic pathway.
We use these cookies to ensure our site functions securely and properly; they are necessary for our services to function and cannot be switched off in our systems. They are usually only set in response to actions made by you which amount to a request for services, such as logging in, using a shopping cart or filling in forms.
You can set your browser to block or alert you about these cookies, but some parts of our services will not work without them. Like the other cookies we use, strictly necessary cookies may be either first-party cookies or third - party cookies.
We use these cookies to remember your settings and preferences. For example, we may use these cookies to remember your language preferences. We use these cookies to collect information about how you interact with our services and to help us measure and improve them.
For example, we may use these cookies to determine if you have interacted with a certain page. We and our advertising partners use these cookies to deliver advertisements, to make them more relevant and meaningful to you, and to track the efficiency of our advertising campaigns, both on our services and on other websites and social media.
Visit our Privacy Policy and Cookie Policy pages to learn more about these topics. Deselect this checkbox to disable cookies related to targeted advertising in your Promega experience. These advertising cookies will no longer be set for this browser and device.
Note: These settings are based on necessary cookies that are placed on your device, and if those cookies are cleared or you access the website from another browser or device, advertising cookies might be reset by default in the future, from which you can opt-out.
Manage Cookies Reject All Cookies Yes, I Accept. Your Account Username Account not found. Email address is unverified. Account is locked. Password Incorrect password. Password reset is required. Account is invalid. Log In. Create an Account.
Contact Customer Service Forgot Password? To protect your privacy, your account has been locked after 6 failed login attempts. Please contact Customer Service to unlock your account.
Enter your username and we'll send a link to reset your password. Username Username not found. Send Email. A password reset email has been sent to the primary email address associated with your account. Current Password Incorrect password. New Password Minimum of 8 characters Uppercase and lowercase letters At least one number.
Password doesn't meet requirements. Password has been used too recently. Confirm New Password Passwords don't match. You have successfully reset your password.
Your password reset link has expired. Please request another reset link. Reset Password. There was an issue resetting your password. There was an issue logging into your account. A verification email has been sent to the primary email address associated with your account.
There was an issue sending the verification email. Name This field is required. Email Address Please enter a valid email address. Username already in use.
Password Password doesn't meet requirements. Minimum of 8 characters Uppercase and lowercase letters At least one number. Confirm Password Passwords don't match.
: Autophagy and autophagy flux quantification| Measuring Autophagic Flux: A Simple Guide to How and Why | D , HeLa eGFP-LC3B cells were either mock-treated or treated with, Chloroquine 10 µM or Rapamycin 1 µM for 4 h and subsequently saponin treated. Article CAS PubMed PubMed Central Google Scholar Betz, C. Leucine signals to mTORC1 via its metabolite acetyl-Coenzyme A. Thus, it is possible to screen whole libraries of proteins for autophagy induction see 3. The autophagosome fuses with an endosome to form an amphisome an acidic late autophagosome Forgot your password? Marino, G. |
| The Autophagy Pathway | One possibility is measuring autophagic flux by traditional western blotting that quantifies a protein of interest in your sample. LC3-II is a protein that is found on all autophagosomes think of LC3-II as a little tag on your trash bag, marked to be delivered to the compost and, once the autophagosome fuses with the lysosome, on autolysosomes. Therefore, LC3-II can be measured to understand changes in autophagosome and autolysosome numbers. However, you soon run into the same problem as before; if there is an increase in LC3-II, is that because there is an increase in the trash or a problem delivering the trash to the compost? To overcome this, researchers compare the levels of LC3-II in two different treatment conditions; untreated samples vs. samples treated with drugs that block the autophagosome delivering the trash to the lysosome, known as lysosomal inhibitors. By blocking the delivery of the trash, and the fusion of autophagosomes with lysosomes to form autolysosomes, this will lead to a build-up of autophagosomes, and therefore LC3-II. To help illustrate this, I have continued the trash bag analogy in Figure 1. You can see in scenario one, with the addition of a lysosomal inhibitor, depicted as a wall, there is an accumulation of trash bags autophagosomes as they cannot deliver the trash to the compost lysosome. This would represent normal autophagy biology. In scenario two, however, there seems to be less of an accumulation of trash bags even with a wall blocking them. This would suggest that there are not as many trash bags being delivered to the compost in scenario two. In the context of the autophagy pathway, this would reflect a decrease in autophagosome production, relative to scenario one. In scenario three there is a large amount of trash bags even without the wall, and the addition of the wall does not seem to change that trash bag number much. This would suggest that there is already something preventing the trash from being delivered to the compost. This analogy can be used to monitor changes in LC3-II levels in the presence or absence of lysosomal inhibitors the wall, in our analogy to determine the state of autophagic flux in your samples of interest. Figure 1: measuring autophagic flux using lysosomal inhibitors. In some experimental setups, it will not be possible to use lysosomal inhibitors. For example, lysosomal inhibitors are highly suitable for in vitro studies i. directly added to cells in a dish. However, what if you wanted to monitor autophagic flux in vivo e. in a whole, intact organ or system, such as a brain slice? In these instances, administering lysosomal inhibitors to a whole organism may not be appropriate. If you are located in the EEA European Economic Area , the United Kingdom, or Switzerland, you can change your settings at any time by clicking Manage Cookies in the footer of our website. Your Account. To protect your privacy, your account will be locked after 6 failed attempts. After that, you will need to contact Customer Service to unlock your account. You have 4 remaining attempts. You have 3 remaining attempts. You have 2 remaining attempts. You have 1 remaining attempt. Contact Customer Service. Forgot Password? Username not found. This field is required. There was an issue with the password reset process. Please try again or contact Customer Service. Log in with Your New Password. You have not verified your email address. A verified email address is required to access the full functionality of your Promega. com account. Resend verification email. Click Here. Autophagy is an important intracellular pathway for degradation and recycling of superfluous components, and for the removal of harmful subcellular materials. We offer an easy-to-use, plate-based autophagy detection assay that quantifies autophagic flux with a simple, bioluminescent signal. The Autophagy LC3 HiBiT Reporter Assay discerns autophagy inducers from inhibitors with a quantitative, plate-reader based readout. This new autophagy detection method uses two reporter-based technologies: HiBiT and HaloTag, enabling plate-reader assays, protein blotting and imaging using a single reporter cell line. The versatile autophagy reporter allows quantitative study of the autophagy pathway, identification of novel modulators of autophagic flux, and confirmation of mechanism of action. Clear all filters. Sort by: Newest Alphabetical A-Z Alphabetical Z-A. Showing 1 of 1 Products. Autophagy is an intracellular degradation process that is implicated in many physiological and pathological conditions. When autophagy is induced, an isolation membrane is formed that encloses part of the cytoplasm and forms a double-membraned vesicle autophagosome. The autophagosome outer membrane fuses with a lysosome to form the autolysosome, degrading the engulfed organelle components along with the inner autophagosome membrane. Mammalian LC3B is an autophagosome marker that is processed by Atg4 at its C-terminus to become LC3-I. LC3-I is mostly cytosolic, but is lipidated through phosphatidylethanolamine PE conjugation to form LC3-II that localizes at both the outer and inner membranes of the autophagosome. Upon autophagosome fusion with the lysosome, the inner-membrane bound LC3-II is degraded by lysosomal proteases. Monitoring the conversion of LC3-I to LC3-II by change in molecular weight is a common method to monitor changes in the autophagic pathway. Imaging LC3B localization in the cell from diffuse in the form of cytostolic LC3-I to concentrated LC3-II in autophagosomes or puncta is an additional method to track changes in autophagy. There are several methods for monitoring autophagosome formation and autophagic flux. However, each method has its limitations, and more than one method should be used to validate autophagic activity. For example, using the number of autophagosomes determined by GFP-LC3 localization is a common method for measuring autophagy activity. However, the results might be misleading because both induction and inhibition of autophagy can lead to increased autophagosome accumulation, depending on where in the pathway the perturbation occurs. The Autophagy LC3 HiBiT Reporter Assay System can be used in three detection modules to unambiguously quantitate autophagic flux. First, the reporter assay measures autophagic flux by monitoring total levels of the LC3-based reporter in a plate-reader based assay. Cells stably expressing the autophagy reporter and treated with stimulators of autophagy will show a decreased luminescent signal. Treatment with inhibitors of autophagy results in a buildup in the level of LC3-based reporter and thus a higher luminescent signal. Second, the HiBiT tag on the LC3 reporter can be used with the Nano-Glo® HiBiT Blotting System to monitor the molecular weight change from LC3-I to LC3-II. This system replaces cumbersome western blots with a faster and simpler protocol. Lysosomal positioning coordinates cellular nutrient responses. Betz, C. Where is mTOR and what is it doing there?. Fedele, A. Chloroquine and bafilomycin A mimic lysosomal storage disorders and impair mTORC1 signalling. Article Google Scholar. Zoncu, R. Li, M. Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 MTORC1 activity. Yu, L. Termination of autophagy and reformation of lysosomes regulated by mTOR. Chen, R. Lynch, C. Role of leucine in the regulation of mTOR by amino acids: Revelations from structure-activity studies. Marino, G. Regulation of autophagy by cytosolic acetyl-coenzyme A. Cell 53 , — Chen, L. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. Berl 81 , — Hariharan, N. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Sebti, S. BAT3 modulates pdependent acetylation of p53 and autophagy-related protein 7 ATG7 during autophagy. USA , — Lee, I. Regulation of autophagy by the p acetyltransferase. Esteves, A. Song, T. Acetylation modulates LC3 stability and cargo recognition. FEBS Lett. Salminen, A. SIRT1: Regulation of longevity via autophagy. Cell Signal. Son, S. Leucine signals to mTORC1 via its metabolite acetyl-Coenzyme A. Pietrocola, F. Spermidine induces autophagy by inhibiting the acetyltransferase EP Cell Death Differ. Download references. We thank Dr. Zhiqin Ji AbbVie for the synthesis of the AbbVie-internal EP inhibitor, Dr. Kenneth Bromberg AbbVie for help and support regarding activities on EPinhibitors, as well as Dr. Janina Ried AbbVie for advice on statistical analysis. Martina P. Clinical Cooperation Unit Translational Radiation Oncology, National Center for Tumor Diseases NCT , Heidelberg University Hospital UKHD and German Cancer Research Center DKFZ , Heidelberg, Germany. Faculty of Biosciences, Heidelberg University, Heidelberg, Germany. Department of Biotechnology, Mannheim University of Applied Sciences, Mannheim, Germany. Centre for Infectious Diseases, Heidelberg University Hospital, Heidelberg, Germany. You can also search for this author in PubMed Google Scholar. and J. performed and analyzed experiments. and V. supervised the project. wrote the manuscript text, M. L and C. prepared figures. All authors reviewed the manuscript. Correspondence to Martina P. Liebl or Viktor Lakics. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Open Access This article is licensed under a Creative Commons Attribution 4. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. Reprints and permissions. Liebl, M. Robust LC3B lipidation analysis by precisely adjusting autophagic flux. Sci Rep 12 , 79 Download citation. Received : 12 July Accepted : 06 December Published : 07 January Anyone you share the following link with will be able to read this content:. Sorry, a shareable link is not currently available for this article. Provided by the Springer Nature SharedIt content-sharing initiative. By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate. Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily. Skip to main content Thank you for visiting nature. nature scientific reports articles article. Download PDF. Subjects Biological techniques Cell biology Macroautophagy. Abstract Autophagic flux can be quantified based on the accumulation of lipidated LC3B in the presence of late-stage autophagy inhibitors. Introduction Modulation of macro-autophagy hereafter referred to as autophagy presents a promising therapeutic strategy in a variety of human diseases, ranging from cancer to neurodegeneration. Results Non-saturating concentrations of BafA result in a wide assay window when measuring LC3-II lipidation Clonal cell lines often show cell-type specific sensitivity towards late-stage autophagy inhibitors and longer than 24 h treatment with these compounds frequently results in acute toxicity and cell death, especially at higher concentrations data not shown. Figure 1. Full size image. Figure 2. Figure 3. Figure 4. Figure 5. Figure 6. Discussion One of the most frequently used methods to detect changes in autophagic flux is the Western blot-based, semiquantitative measurement of LC3-II levels upon stimulation or inhibition of macro-autophagy. Autophagy LC3 HiBiT reporter assay HiBiT-LC3B expressing HEK cells and the assay system were obtained from PROMEGA. Ethical approval This study does not contain any studies with human or animal subjects performed by any of the authors. References Heitman, J. Article ADS CAS PubMed Google Scholar Chiu, M. Article ADS CAS PubMed PubMed Central Google Scholar Sabers, C. Article CAS PubMed Google Scholar Hosokawa, N. Article CAS PubMed PubMed Central Google Scholar Ganley, I. Article CAS PubMed PubMed Central Google Scholar Kim, Y. Article CAS PubMed Google Scholar Pena-Llopis, S. Article CAS PubMed PubMed Central Google Scholar Ravikumar, B. Article CAS PubMed Google Scholar Webb, J. Article CAS PubMed Google Scholar Ravikumar, B. Article CAS PubMed Google Scholar Berger, Z. Article CAS PubMed Google Scholar Ozcelik, S. Article ADS CAS PubMed PubMed Central Google Scholar Bjedov, I. Article CAS PubMed PubMed Central Google Scholar Ha, C. Article CAS PubMed Google Scholar Miller, R. Article CAS PubMed PubMed Central Google Scholar Lamming, D. Article CAS PubMed PubMed Central Google Scholar Arriola Apelo, S. Article CAS Google Scholar Werner, G. Article PubMed Google Scholar Crider, B. Article CAS PubMed Google Scholar Zhang, J. Article CAS PubMed Google Scholar Homewood, C. Article ADS CAS PubMed Google Scholar Seglen, P. Article CAS PubMed Google Scholar Poole, B. Article CAS PubMed Google Scholar Mauthe, M. CAS PubMed PubMed Central Google Scholar Bjorkoy, G. Article PubMed PubMed Central CAS Google Scholar Pankiv, S. Article CAS PubMed Google Scholar Lamark, T. Article PubMed Google Scholar Itakura, E. CAS PubMed PubMed Central Google Scholar Komatsu, M. Article CAS PubMed Google Scholar Korolchuk, V. Article PubMed Google Scholar Michaelides, M. Article PubMed Google Scholar Dixon, A. Article CAS PubMed Google Scholar Florey, O. Article CAS PubMed PubMed Central Google Scholar Jacquin, E. Article CAS PubMed PubMed Central Google Scholar Sanjuan, M. Article ADS CAS PubMed Google Scholar Heckmann, B. Article CAS PubMed PubMed Central Google Scholar Gao, Y. Article CAS PubMed PubMed Central Google Scholar Yamamoto, A. Article CAS PubMed Google Scholar Kawai, A. Article CAS PubMed Google Scholar Klionsky, D. Article CAS PubMed Google Scholar Redmann, M. Article CAS PubMed Google Scholar Settembre, C. Article ADS CAS PubMed PubMed Central Google Scholar Saucedo, L. Article CAS PubMed Google Scholar Sancak, Y. Article CAS PubMed PubMed Central Google Scholar Korolchuk, V. Article CAS PubMed PubMed Central Google Scholar Betz, C. Article CAS PubMed PubMed Central Google Scholar Fedele, A. Article Google Scholar Zoncu, R. Article ADS CAS PubMed PubMed Central Google Scholar Li, M. Article CAS PubMed PubMed Central Google Scholar Yu, L. Article ADS CAS PubMed PubMed Central Google Scholar Chen, R. Article CAS PubMed PubMed Central Google Scholar Lynch, C. Article CAS PubMed Google Scholar Marino, G. Article CAS PubMed Google Scholar Chen, L. Article CAS Google Scholar Hariharan, N. Article CAS PubMed PubMed Central Google Scholar Sebti, S. Article ADS CAS PubMed PubMed Central Google Scholar Lee, I. Article CAS PubMed PubMed Central Google Scholar Esteves, A. Article CAS PubMed Google Scholar Song, T. Article CAS PubMed Google Scholar Salminen, A. Article CAS PubMed Google Scholar Son, S. Article CAS PubMed PubMed Central Google Scholar Pietrocola, F. Article CAS PubMed Google Scholar Download references. Acknowledgements We thank Dr. Meister Faculty of Biosciences, Heidelberg University, Heidelberg, Germany Sarah C. Meister Department of Biotechnology, Mannheim University of Applied Sciences, Mannheim, Germany Lisa Frey Mikrogen GmbH, Neuried, Germany Kristina Hendrich Centre for Infectious Diseases, Heidelberg University Hospital, Heidelberg, Germany Anja Klemmer Authors Martina P. Liebl View author publications. View author publications. Ethics declarations Competing interests The authors declare no competing interests. Additional information Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Supplementary Information. Rights and permissions Open Access This article is licensed under a Creative Commons Attribution 4. About this article. Cite this article Liebl, M. Copy to clipboard. This article is cited by Glial senescence enhances α-synuclein pathology owing to its insufficient clearance caused by autophagy dysfunction Bin Hong Yosuke Ohtake Toshihide Yamashita Cell Death Discovery Deficiency of RAB39B Activates ER Stress-Induced Pro-apoptotic Pathway and Causes Mitochondrial Dysfunction and Oxidative Stress in Dopaminergic Neurons by Impairing Autophagy and Upregulating α-Synuclein Ching-Chi Chiu Yi-Hsin Weng Hung-Li Wang Molecular Neurobiology Comments By submitting a comment you agree to abide by our Terms and Community Guidelines. |
| Shop all Autophagy Detection | Intact cells were gated in FSC-A vs SSC-A and single cells gated in FSC-H vs FSC-A. Stringent gating strategies to exclude debris or dead cells that exhibit autofluorescence were applied. Sequencing was performed on the Illumina Genome Analyzer in the GeneCenter sequencing facility LAFUGA. Obtained sequences were processed with the Trim Galore! toolkit to remove adapter sequences and reads with PHRED scores below 30 as previously described 39 , Default settings were used to process the raw reads. Combined from three independent experiments, 6. To align the reads to the GeCKO 2. The raw sequencing data were deposited in GEO ID: GSE Data were collected at least in triplicates for all flow cytometry-based approaches and with at least 30 replicates for fluorescence confocal microscopy-based approaches. For the relative comparison of changes in autophagic flux, statistical differences were assessed by one-way ANOVA or unpaired t-test as indicated. A p-value below 0. To evaluate the quality of the method for high-throughput screening, we calculated the Z-factor as described previously A Z-factor above 0 indicates a method suitable for high-throughput approaches, 1 represents the maximum quality reachable. A detailed list of materials is provided in Table 1. For the construction of the cell lines use low passage numbers. For accurate autophagy measurements, make sure that the cells never overgrew, even during the construction of the cell lines, discard any flasks which are too dense. Cell lines should be discarded once the passage number is higher than 20 assuming a biweekly split-cycle. Amplify a fused eGFP-LC3B ORF from the pBabe-mCherry-eGFP-LC3B template and insert it into the pBob-eGFP vector, replacing the existing eGFP ORF:. Digest the amplified insert ca. Digest the pBob-eGFP vector using BamHI and PmeI, purify on an agarose gel and recover the DNA Silica Bead DNA Gel Extraction Kit, Thermo Scientific to create the backbone for ligation. Optional : An internal ribosomal entry site IRES element can be inserted after the eGFP-LC3B fusion ORF to allow the expression of an antibiotic resistance protein. To avoid selection using antibiotics and stressing the autophagy reporter cells, we decided to omit this. Mix B: Opti-MEM µl per well of the 6-well plate, Gibco and plasmid DNA for the amounts see Table 4. Repeat if no fluorescence is observed. Harvest the SN 48 h post transfection and clear it by centrifugation 2, g, 10 min, 4 °C and filtering Millex-HA Filter, 0. HEKT, HeLa with µl of the SN harvested in step 1. Two days post-transduction, detach the cells using Trypsin 0. Filter through round-bottom tubes with a cell strainer cap 5 ml, Falcon. Sort samples on a FACS sorter e. BD FACSAria III, using a micron ceramic nozzle. The flow rate was kept below 2, and the purity setting was set to a single cell. Sort one cell per well F-bottom well. Important: The nozzle size needs to be larger than the cell passing through it. Avoid crushing or stressing the cells too much. Select eGFP-LC3B cells, with moderate eGFP MFIs Fig. Construction of autophagy reporter cell lines. A Schematic overview. After random genomic integration of an eGFP-LC3B expressing cassettes into target cells by lentiviral delivery, single clones are isolated via FACS sorting and grown into clonal cell lines. B FACS sorting gating strategy for eGFP-LC3B-lentivirus transduced HeLa cells. Viable cell clones with intermediate eGFP-LC3B expression were chosen blue. C Exemplary images of HeLa single cell clones expressing eGFP-LC3B in well plates. Left panels: brightfield image and corresponding eGFP fluorescence green of a single clone after 1 week. Right panel, brightfield image and corresponding eGFP fluorescence green after 3 weeks, before transfer into larger wells. Size marker, µm. D Detection of eGFP-LC3B expression in isolated clonal HEKT, HeLa, Jurkat and THP-1 cell lines, comparing them to their respective parental cell line. Immunoblotting of whole cell lysates using anti-GFP, anti-LC3B and anti-β-actin antibodies. Uncropped western blots in Supplementary Fig. Important : Some cell lines do not grow efficiently from single clones. Avoid isolating cell lines expressing too much or no eGFP-LC3B. Approximately 3—6 weeks post sorting, clones can be transferred from half-confluent wells directly into well plates. Avoid overgrowing in wells or at any step. eGFP-LC3B expression and fluorescence can be monitored using a fluorescence microscope and clones with little to no detectable fluorescence are discarded. Clones with clearly visible aggregations of eGFP-LC3B in the cytoplasm are also discarded. Generate whole-cell lysates of the cell line as described in the short Methods section , and immunoblot with anti-GFP antibody and anti-LC3B antibody. An additional band at around 50 kDa, representing the fusion protein, should appear in the stable cell lines compared to their parental cell lines. Note : In some cell lines an additional band at the height of eGFP ca. We have not observed any negative impact of this additional eGFP band on the sensitivity and responsiveness of the respective cell lines. Cell lines with further degradation products should be discarded. Take confocal images and analyze the number of autophagosomes upon treatment with positive controls such as Rapamycin 1 µM or Chloroquine 10 µM for 4 h. Follow the protocol for high-throughput autophagy quantification to verify that the cell line is suitable for the approach and contains enough eGFP-LC3B for cytoplasmic washout and subsequent analysis. Use treatment with Rapamycin 1 µM or Chloroquine 10 µM for 4 h and compare to cells treated with the carrier e. Dimethyl sulfoxide DMSO or water. Important : The level of expressed eGFP-LC3B impacts the sensitivity of the assay. Generally, we have found that a medium amount of eGFP-LC3B seems to be optimal. Too low eGFP-LC3B expression causes a loss of detectable eGFP-LC3B fluorescence after washout. Too high eGFP-LC3B causes a high baseline of autophagosome associated eGFP-LC3B, as well as occasionally aggregates observed in confocal microscopy. Produce frozen stocks as early as possible to preserve the generated cell line at a passage number as low as possible. A detailed list of materials is provided in Table 5. Grow cells in either F-bottom adherent cell types or V-bottom suspension cell types well plates. Seeding of 50, cells per well for adherent cells and , cells per well for non-adherent cells is recommended, 18 h before a 4-h assay. Important : At the time of harvest, the cells should be approximately confluent, and kept in fully supplemented medium which is still at optimal pH. Any stress on the cells impacts autophagy and thus may bias the assay. Consider appropriate negative and positive controls. Established drugs like Rapamycin, Chloroquine, or Bafilomycin A1 are recommended as controls. Negative controls should be treated the same way as the samples, e. add the carrier for a drug in the same concentration to the mock SN. Screening results have to be confirmed using orthogonal methods for assessing autophagy levels. Important : Proceed with the harvesting as quickly as possible until the cells are in saponin-containing PBS step 2. However, at a loss of cell count. We have found that washing twice is sufficient for most applications, and cell numbers stay within a reasonable range. Use one of three different subsequent treatments see Fig. Important : Cells disintegrate rapidly if they are not fixed after saponin treatment, however, the cells are good for FACS analysis for up to 3 h following the treatment if extensively washed. Important: Fixed cells can be stored in the fridge 4 °C for analysis up to a week. But fluorescence of the cells wanes over time. Analyze as early as possible. Proceed to flow cytometry. Note that saponin-treated cells appear smaller than non-treated cells and have less eGFP content, as well as less autofluorescence, see Fig. Important : Quantification using less than 10, cells still is possible, however, the inherent heterogeneity of autophagy, even within cell populations derived from a single clone, may result in higher deviations between biological replicates. Extract the MFI of eGFP-LC3B from all samples. For background correction, the MFI measured in mock conditions is subtracted from the MFI of treated samples see Flow Cytometry and Cell Sorting. A detailed list of materials is provided in Table 6. Important: All hits in screening approaches have to be verified using orthogonal methods to assess autophagy for a comprehensive overview of available methods see Seed eGFP-LC3B reporter cells in well plates, 50, cells per well in F-bottom plates for adherent cells, and , cells per well in V-bottom plates for non-adherent cells. The total amount of medium should be 90 µl fully supplemented medium e. Seed non-adherent cells directly before applying the treatment. Add 10 µl of treatment solution to the cells. Several examples of drug treatment were used to verify the generated cell lines. As a control, use the same amount of carrier liquid e. water or DMSO. Important : Do not exchange the medium before treatment. Do not vary the volume between treatments and do not add additional medium, FBS, or salt beyond mM. Any stress may cause changes in autophagic flux and obscure effects of drugs, thus treat your cells carefully. For our exemplary experiments Fig. Rapamycin InSolution Rapamycin, Merck , which induces autophagy by inhibiting the mTORC1 complex 6 , 43 , was used in a range of 2 µM to 15 nM Fig. Chloroquine Chloroquine diphosphate, Santa Cruz Biotechnology , which blocks the turnover of autophagosomes 44 , 45 , 46 , was used in a range of 20 µM to nM Fig. Bafilomycin A1 Santa Cruz Biotechnology , which blocks the turnover of autophagosomes 47 , 48 , was used in a range of nM to 5 nM Fig. Optionally : Cells can be treated longer or shorter, we have observed that a time frame between 2 and 4 h is optimal for most drug-based applications. However, drugs that are expected to block autophagic flux may be kept on the cells for longer to measure the accumulation of vesicles from basal autophagy. Please note that especially blocking autophagy decreases the viability of the cells after 4—6 h. Seed cells in 90 µl fully supplemented medium e. Infect cells by addition of 10 µl virus-containing solution or mock carrier with an appropriate MOI at different time points, e. as indicated in Fig. if the virus was produced in Vero cells, add an equal amount SN from non-infected Vero cells to the mock wells. Alternatively, use the buffer the virus is stored in, e. after purification. Important : Discard virus-containing SN according to your biosafety protocols. Keep in mind that some viruses like EMCV are not inactivated by saponin-treatment. Transfect vectors coding the proteins of interest 18 h post-seeding and the empty vector control using PEI and a maximum of 0. Mix A: Mix Opti-MEM 10 µl per well of the well plate and PEI 2 µl PEI per µg DNA and incubate for 5 min at RT. Mix B: Mix Opti-MEM 10 µl per well of the well plate and DNA 0. Important : Transfection induces autophagy. We have tested a few transfection reagents and found that PEI and calcium phosphate transfection induce the lowest amount of autophagy as opposed to commercially available transfection reagents. Thus, pre-test your transfection method. Alternatively, transduction can be used, however, gene expression is lower and an autophagic response is still induced albeit to a lesser degree. Important : This procedure was modified from Joung et al Please consult the original publication for a comprehensive protocol. Jurkat eGFP-LC3B with an MOI of 0. Determine the initial cell number for the protocol. Take up the cells in 10 ml 1xPBS and filter through a round-bottom tube with a cell strainer cap 5 ml, Falcon. Discard the medium autophagy inducing cells. Adjust the flow rate and the concentration of the treated sample to the highest possible flow rates. Use the Zymo Research Quick-gDNA MidiPrep Kit to extract the genomic DNA from the cells according to the manufacturer's protocol. Set up 10 PCR reactions according to Tables 7 , 8 , and 9. Proceed with next-generation-sequencing. Highly robust quantification of autophagy can be achieved by measuring the amount of LC3B-positive vesicles, a hallmark of autophagy, using eGFP-LC3B expressing reporter cells 24 , To this end, we constructed autophagy reporter cell lines stably expressing eGFP-LC3B from a genomically integrated, CMV-promoter controlled, expression cassette. Third generation lentiviral particles harboring the expression cassette were generated and target cell lines transduced. The cell lines include cell lines HeLa classically used for autophagy research, easy-to-transfect cell lines HEKT and cells of the immune system like Monocyte-like and T cells THP-1 and Jurkat Fig. Following transduction, single-cell clones were sorted from a pool of medium level eGFP-LC3B expressing cells exemplarily shown for HeLa eGFP-LC3B cells in Fig. These single-cell clones were grown into clonal cell lines and eGFP-LC3B fluorescence monitored by fluorescence microscopy Fig. Unchanged p62 levels between parental and stable cell lines suggest that autophagic flux was not significantly altered by the expression of eGFP-LC3B Fig. Finally, expression and size of the fusion protein eGFP-LC3B ca. All reporter cell lines Fig. To confirm that the reporter cell lines respond to stimulation and blockage of autophagy, they were treated with different drugs that induce Rapamyicin or block autophagic flux Chloroquine, Bafilomycin A1. Chloroquine and Bafilomycin A1 treatment led to an accumulation of perinuclear autophagosomes Fig. The pixel area of eGFP-LC3B, which correlates with autophagy levels, was quantified using semi-automated analysis Fig. Confocal images were taken and randomly selected single cells extracted. Aided by an ImageJ macro 50 , automatic thresholding, and particle counting resulted in the pixel area of autophagosomes Fig. Western blotting confirmed that p62 accumulates in the reporter cell lines upon Bafilomycin A1 treatment, demonstrating functional autophagy Fig. Taken together, the generation of eGFP-LC3B expressing autophagy reporter cell lines derived from HeLa, HEKT, Jurkat, and THP-1 cells was successful. A Schematic overview of the method. Using ImageJ, the area and the count of eGFP-LC3B puncta in the cells can be quantified to measure autophagy. B Exemplary confocal images of eGFP-LC3B green expressing HeLa, HEKT, Jurkat, and THP-1 cells. The cells were either mock-treated or treated with Rapamycin 1 µM , Chloroquine 10 µM , or Bafilomycin A1 2. Nuclei, DAPI blue. Size marker, 10 µm. C Quantification of the eGFP area per cell of HeLa eGFP-LC3B cells, HEKT eGFP-LC3B, Jurkat eGFP-LC3B, or THP-1 eGFP-LC3B either mock-treated or treated with Rapamycin 1 µM , Chloroquine 10 µM , or Bafilomycin A1 2. For high-throughput applications, an efficient system to quantify LC3B-positive autophagosomes is desirable. To measure autophagy using flow cytometry, eGFP-LC3B-II decorated autophagosomes have to be separated from cytosolic eGFP-LC3B-I. Thus, eGFP-LC3B expressing cells were permeabilized with 0. As determined by flow cytometry, the cells decreased in size and granularity Fig. Complete permeabilization as indicated by cell size is reached after 10 min Fig. Furthermore, after successful removal of cytoplasmic eGFP-LC3B-I, the mean eGFP fluorescence levels are drastically reduced Fig. S1 D , representing only fluorescence of autophagosome-bound eGFP-LC3B-II Fig. Therefore, treatment with saponin allows quantification of changes in autophagosome numbers, which are not visible in non-permeabilized cells Fig. The washout procedure is very robust, and only minor variations are observed in the treatment Fig. It is possible to fix the cells to preserve the eGFP-LC3B signal after saponin permeabilization for longer storage using two different methods: PFA and MeOH. Compared to non-fixed cells, the signal in PFA-fixed cells was well preserved, MeOH fixation, however, caused a slight drop of the absolute eGFP-LC3B signal Fig. However, as the differences between differently treated cells were preserved, both fixation methods were suitable. Thus, isolation of autophagosome-bound eGFP-LC3B and its detection using flow cytometry was successful and can be used to reveal changes in autophagy levels. A , Schematic overview of the method. eGFP-LC3B expressing cell lines are permeabilized with 0. Using FACS, the amount of membrane-bound eGFP-LC3B per cell is quantified as eGFP mean fluorescent intensity MFI. B , Forward and side scatter of HeLa eGFP-LC3B cells in flow cytometry before blue and after saponin treatment red top panel. eGFP-LC3B fluorescence is decreased after saponin treatment of HeLa eGFP-LC3B cells bottom panel. C , Total eGFP-LC3B MFI of HeLa eGFP-LC3B cells, either mock-treated or treated with Rapamycin 2 µM, 4 h top panel. eGFP-LC3B MFI of HeLa eGFP-LC3B cells, either mock-treated or treated with Rapamycin 2 µM, 4 h after saponin permeabilization and cytoplasmic washout of the cells bottom panel. D , HeLa eGFP-LC3B cells were either mock-treated or treated with, Chloroquine 10 µM or Rapamycin 1 µM for 4 h and subsequently saponin treated. eGFP MFIs were quantified, either in non-fixed green , methanol fixed blue , or paraformaldehyde PFA fixed cells red. The left panel shows the raw MFI, the right panel shows the background adjusted values. Statistical significance was assessed using unpaired t-tests C or one-way ANOVA D. Drug screenings, as exemplified for the known autophagy manipulating drugs Rapamycin, Chloroquine, and Bafilomycin A1, can be readily performed Fig. Dotted red lines represent twice the standard deviation of the mock control to illustrate the sensitivity of the approach. For all treatments, Z-factors ranging between 0. A Z-factor above 0 indicates high robustness of the high-throughput approach 42 , suggesting that the method is robust enough for high-throughput applications. As a proof-of-principle whether our approach can be used to detect novel autophagy modulating compounds, we assessed the impact of 18 different human amino acids on eGFP-LC3B levels. Our results indicate that whereas amino acids like cysteine, isoleucine, asparagine, serine, valine or threonine may induce autophagy, others like arginine, tyrosine or glycine slightly reduce autophagic flux Fig. This is in accordance with previous reports that indicate a role of amino acids in the modulation of autophagy 51 , 52 , Taken together, all cell lines that were generated responded accurately, robustly, and sensitively to drug treatment. Thus, this approach is suitable for high-throughput quantification of autophagy to discover novel compounds that modulate autophagy see 3. Analysis of autophagy-modulating compounds using eGFP-LC3B expressing cell lines. Cells were either mock-treated or treated with Rapamycin 2 µM—15 nM; left panel , Chloroquine 20 µM— nM; middle panel , or Bafilomycin A1 nM—5 nM, right panel for 4 h. Z-factors are indicated next to the respective diagram. The red dotted line indicates the double standard deviation of the mock control. Whereas influenza A virus IAV infection significantly increased autophagosome levels in epithelial cells already after 6 h 10 , 54 , 55 , measles virus MeV infection induced high numbers of autophagosomes at late time points 18 h, 24 h, 48 h 56 , In monocyte-like cells, both IAV and MeV infection induced autophagy at early time points Fig. Infection with encephalomyocarditis virus EMCV rapidly induced high levels of autophagosomes 10 , 58 Fig. In summary, time-dependent changes in autophagy induced by viral infection can be accurately monitored on a small scale well using our system. Furthermore, samples with higher biosafety levels above BSL1 can be easily processed and fixed, and then safely analyzed in BSL1 conditions see 3. A , HeLa GL cells were infected with influenza A virus IAV, left panel, MOI 5 , encephalomyocarditis virus EMCV, middle panel, MOI 10 or measles virus MeV, right panel, MOI 5. Cells were harvested, saponin treated, fixed, and the eGFP-LC3B fluorescence analyzed by flow cytometry at the indicated time points post infection. Treatment with Chloroquine 1 µM, 4 h and Rapamycin 1 µM, 4 h served as controls. B , HEKT GL cells were transiently transfected with an empty vector or a TRIMFLAG expressing construct. Cells were saponin treated, fixed, and stained with anti-FLAG antibodies APC left and middle panel. Each dot is derived from 6 individual sgRNAs targeting the same gene. The dotted red line indicates an equal abundance of sgRNA counts in both populations. Statistical significance was assessed using unpaired t-test A or one-way ANOVA B. Uncropped agarose gel in Supplementary Fig. Autophagy induction by protein transfection into HEKT eGFP-LC3B cells can be rapidly assessed as exemplified by inducing autophagy with TRIM32 overexpression 59 Fig. S2 B in HeLa reporter cells. Thus, it is possible to screen whole libraries of proteins for autophagy induction see 3. S2 C and D. We successfully extracted genomic DNA from Jurkat eGFP-LC3B cells after processing and MeOH fixation as described in our basic protocol. The samples were processed and fixed with MeOH according to our basic protocol. The sgRNA cassette was amplified by PCR Fig. From a total , individual sgRNAs in the original GeCKO library, we could obtain sequences for S2 F , demonstrating that complexity was retained. Knockout of components of the autophagic machinery should lower autophagosome levels. This confirms that the method is able to identify components of the autophagic machinery. A wide variety of methods to reliably quantify autophagy is currently available for a comprehensive review see These methods include visualization of autophagosomes by electron microscopy, monitoring of degradation of targets of autophagy such as p62 using western blotting, processing of endogenous LC3B, and visualization of LC3B puncta using eGFP-LC3B and confocal fluorescence microscopy. Advanced imaging methods using automated image processing reduce the manual labor required for confocal image acquisition and analysis Several currently used methods to quantify autophagy rely on monitoring a hallmark of autophagy induction, the lipidation and translocation of eGFP-LC3B to autophagosomal membranes. However, most of these methods are not applicable for high-throughput approaches. Compared to classical methods for monitoring autophagy, like western blotting, this system is less labor-intensive, faster, and allows the quantification of large numbers of cells 10, vs 50— at once, allowing extraction of robust means of autophagy levels but also visualization of the heterogeneity of cell systems. Therefore, this system is well fitted for approaches that measure the mean induction of autophagy by e. Proof-of-principle assays revealed that novel autophagy modulating compounds like e. human amino acids can be readily identified. Alteration in autophagy levels due to viral infections can be assessed over time. Our data further reveal that overexpression approaches monitoring the autophagy response of individual cells are possible. Proteins above a size threshold of approximately 50 kDa can be easily co-stained, allowing e. overexpression screenings. To avoid washout of the co-stained protein, it may be anchored via a tag e. GPI anchor to a membrane and thus be retained in the cell sgRNA amplicons were extracted and analyzed by NGS. Taken together, we demonstrate that our protocol is suitable for a wide range of high-throughput approaches that can be applied to answer various scientific problems, ranging from pathway analysis and key factor identification to drug discovery. Autophagy is a highly dynamic process, relying on the complex turnover and activation of signaling cascades. While the reporter cell lines allow fast processing of samples and quantification of autophagy, its use is limited to genetically modified cell lines. The consistent presence of the reporter is mandatory and transiently transfected cell lines do not provide adequate stability of the signal. Thus, the screen should be complemented with monitoring the endogenous LC3B status in primary cells to support conclusions indicated by initial screening methods 24 , One major issue of all systems relying on the processing of LC3B is that both de novo induction of autophagy and blockage of autophagic flux increases the amount of autophagosomes or processed LC3B at a given time point. Thus, a rigorous secondary assessment of hits obtained in the primary screen has to take place for a comprehensive review of methods see Complementary assays to monitor autophagy, that do not rely on LC3B are mandatory to properly assess the status of autophagy in a cell. Alternatively, other cellular proteins targeted by autophagy like NBR1 64 or cGAS 65 can be used as indicators of autophagic degradation. Autophagy-like cellular processes such as LC3-associated phagocytosis 66 or viruses may use LC3B and redirect it to membranes, thus giving rise to false-positive signals in our flow cytometry assay and other methods that measure autophagy using LC3B processing or localization 3 , 54 , In general, for the majority of non-canonical autophagic processes, degradation of p62 is not observed. For example, while LC3B associated phagocytosis is dependent on most core machinery factors, it is independent of e. ATG14L, which is required for canonical autophagy 3 , Besides relying on eGFP-LC3B, other high-throughput methods to quantify autophagy are available. Cell lines expressing the double fluorescence reporter fusion to LC3B mRFP-eGFP-LC3B allow rapid quantification of autophagy without cytoplasmic washout Upon induction of autophagy, eGFP fluorescence is lost in the acidic environment of autophagolysosomes, thus the ratio between eGFP and mRFP fluorescence decreases. Occasionally, eGFP fluorescence is not completely quenched by the acidic pH, resulting in remaining signal, thus alternative fluorophores have been proposed Still, recently novel autophagy regulating factors like TMEM41B 30 were discovered using mRFP-eGFP-LC3B reporter constructs Recently, another sophisticated approach to quantify autophagic flux using flow cytometry was described Using a reporter construct expressing a fusion protein of GFP, LC3, RFP, and a non-cleavable variant of LC3B, LC3BΔG, this system allows, similarly to the double-labeled LC3B, monitoring of autophagy based on the ratio between GFP and RFP fluorescence. Upon autophagy induction, the protease ATG4 is activated and cleaves the fusion protein into GFP-LC3and RFP-LC3BΔG. Eventually, only GFP-LC3B can be incorporated into autophagosomes and is degraded. Thus, upon induction of autophagy, the ratio between GFP and RFP fluorescence decreases. This system has allowed the identification of several novel autophagy modulators However, recombination between the two LC3B ORFs, especially during lentiviral driven applications like CRISPR screens, may render the system inactive. Instead of assessing the number of autophagosomes to quantify autophagy, the consequences of the induction of autophagic flux can also be monitored by examining p62 levels. However, upon induction of autophagy, transcriptional upregulation of p62 has been observed in some instances. Furthermore, longer assay times are needed to allow the degradation to proceed enough to detect decreased levels of p Lysosomal dyes like lysotracker may be used to stain lysosomes or autophagolysosomes and even to monitor changes in pH 24 , 71 , 72 , Thus, systems based on lysosomal dyes are rarely used to monitor autophagy. Taken together, using eGFP-LC3B as a reporter for quantification of autophagy is currently still the most established and advantageous system for quantifying autophagy. It avoids convoluted reporter systems and thus directly quantifies autophagosomes. Our method applies this established tool for high-throughput approaches. Accurate and robust quantification of autophagosomes by measuring the mean fluorescence intensity of membrane-bound eGFP-LC3B is a powerful high-throughput tool to study autophagy. Our protocol is designed to easily approach e. The setup is easy to adopt and provides a robust, flexible, and rapid readout. However, screening results have to be confirmed using orthogonal methods for assessing autophagy levels. Yin, Z. Autophagy: machinery and regulation. Cell 3 , — PubMed PubMed Central Google Scholar. Feng, Y. The machinery of macroautophagy. Cell Res. CAS PubMed Google Scholar. Galluzzi, L. et al. Molecular definitions of autophagy and related processes. EMBO J. CAS PubMed PubMed Central Google Scholar. Zaffagnini, G. Mechanisms of selective autophagy. Moreau, K. Cytoprotective roles for autophagy. Cell Biol. He, C. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43 , 67—93 Gomes, L. Autophagy in antimicrobial immunity. Cell 54 , — Choi, Y. Autophagy during viral infection: a double-edged sword. Sparrer, K. TRIM proteins: new players in virus-induced autophagy. PLoS Pathog. Google Scholar. TRIM23 mediates virus-induced autophagy via activation of TBK1. Crotzer, V. Autophagy and its role in MHC-mediated antigen presentation. Deretic, V. Autophagy balances inflammation in innate immunity. Autophagy 14 , — Levine, B. Autophagy in the pathogenesis of disease. Cell , 27—42 Apel, A. Autophagy: a double-edged sword in oncology. Cancer , — Chen, W. Autophagy: a double-edged sword for neuronal survival after cerebral ischemia. Neural Regen. Henderson, P. Cells 1 , — Onorati, A. Targeting autophagy in cancer. PubMed Google Scholar. Thorburn, A. Autophagy and cancer therapy. Autophagy Cancer , — Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Drug Discov. Kabeya, Y. Erratum: LC3, a mammalian homolog of yeast Apg8p, is localized in autophagosome membranes after processing EMBO Journal 19 — CAS Google Scholar. Geng, J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep. Ohsumi, Y. Molecular dissection of autophagy: two ubiquitin-like systems. Tanida, I. LC3 conjugation system in mammalian autophagy. Klionsky, D. Guidelines for the use and interpretation of assays for monitoring autophagy 3rd edition. Autophagy 12 , 1— A comprehensive glossary of autophagy-related molecules and processes 2nd edition. Autophagy 7 , — Kaizuka, T. An autophagic flux probe that releases an internal control. Cell 64 , — Pietrocola, F. Spermidine induces autophagy by inhibiting the acetyltransferase EP Cell Death Differ. Dejesus, R. Elife 5 , 1—16 Strohecker, A. Oncogene 34 , — Shoemaker, C. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. Jia, R. Negative regulation of autophagy by uba6-birc6—mediated ubiquitination of lc3. eLife 8 , e Petcherski, A. An autophagy modifier screen identifies small molecules capable of reducing autophagosome accumulation in a model of CLN3-mediated neurodegeneration. This would suggest that there is already something preventing the trash from being delivered to the compost. This analogy can be used to monitor changes in LC3-II levels in the presence or absence of lysosomal inhibitors the wall, in our analogy to determine the state of autophagic flux in your samples of interest. Figure 1: measuring autophagic flux using lysosomal inhibitors. In some experimental setups, it will not be possible to use lysosomal inhibitors. For example, lysosomal inhibitors are highly suitable for in vitro studies i. directly added to cells in a dish. However, what if you wanted to monitor autophagic flux in vivo e. in a whole, intact organ or system, such as a brain slice? In these instances, administering lysosomal inhibitors to a whole organism may not be appropriate. An alternative is to use fluorescent proteins to monitor the different parts of the autophagy pathway. Some fluorescent proteins, such as green fluorescent protein GFP , cannot be visualized when they are in an acidic environment such as the lysosome. We can take advantage of this by tagging the LC3 protein a complex of which LC3-II is a part with both GFP, which is sensitive to acidic environments, as well as red fluorescent protein RFP , which is not sensitive to acidic environments and thus monitor the movement of LC3 through the autophagy pathway. When LC3 is on the autophagosome, which is less acidic, both GFP and RFP can be visualized and therefore will appear yellow. However, once the trash has been delivered to the lysosome, the GFP signal will be lost and LC3 will appear red. Thus, monitoring the yellow vs. red signal allows researchers to assess the progress of LC3 through the autophagy pathway and, therefore, autophagic flux. intracranial within the skull injection for expression in the brain. The methods used for measuring autophagic flux are continually growing and improving. Therefore, it is important to have up to date guidelines [1, 2] at hand as well as tips and tricks for optimizations and troubleshooting. You must be logged in to post a comment. This site uses Akismet to reduce spam. Learn how your comment data is processed. Forgot your password? Lost your password? Please enter your email address. You will receive mail with link to set new password. Share this to your network:. X Facebook LinkedIn. Written by Rebecca Wallings. Leave a Comment Cancel Reply You must be logged in to post a comment. Sign in. Remember me. Log in. Reset password. |
| An improved method for high-throughput quantification of autophagy in mammalian cells | About this autophagj. Avoid isolating Goji Berry Supplements lines expressing too much or Antioxidant-rich foods eGFP-LC3B. Close You've successfully Promoting heart wellness out. To align the aitophagy to the GeCKO 2. They count them and see that there is an increase in the number of autophagosomes. The versatile autophagy reporter allows quantitative study of the autophagy pathway, identification of novel modulators of autophagic flux, and confirmation of mechanism of action. |

Welcher bemerkenswert topic