Autophagy and ER stress -
Macroautophagy and microautophagy both carry out the nonselective degradation of proteins, lipids, and organelles [ 50 , 51 ]. The mechanism of autophagy is a complex process that can be categorized into multiple steps.
It involves the formation of double-membrane vesicles containing cellular and external malformed proteins. Long-lived proteins can be induced autophagy, which are ensued by cargo recognition and packaging, an extension of the phagophore membrane, and closure to form the complete autophagosome.
Fusion of the autophagosome with the lysosome occurs, which leads to the derogation of the autophagosomal contents, and the breakdown products are finally eliminated [ 54 , 55 , 56 ].
The initiation of autophagy can be observed by TEM transmission electron microscopy during the expansion of phagophore and autophagosome. The induction of autophagy, vesicle nucleation, and formation of autophagosomes are regulated by the proteins named as autophagy-related genes ATGs [ 50 ].
They are highly conserved genes and were originally discovered in yeasts. Mammalian orthologs of the ATGs have also been discovered [ 57 ]. Autophagy induction is controlled at the molecular level by the multiprotein complex of unclike autophagy-activating kinase 1 ULK1, the mammalian homolog of yeast Atg1 , ATG13, ATGa, and RB1 inducible coiled coil 1 RB1CC1, also known as FIP [ 58 , 59 ].
The c-Jun protein kinase JNK1 and death-associated protein kinase DAPK phosphorylate BCL2 and are positive regulators involved in the induction of autophagy [ 65 , 66 ]. The elongation or obstruction of phagophore depends on two diverse ubiquitin-like protein conjugation reactions [ 67 , 68 ].
The first pathway involves the covalent conjugation reaction of ATG12 to ATG5, with the assist of the E1-like enzyme ATG7 and the E2-like enzyme ATG The second pathway includes the ubiquitin-like system, which plays a role in the conjugation to phosphatidylethanolamine PE lipid and glycine residue of the yeast ATG8 LC3 in the mammal , and is processed by the cysteine protease ATG4 and then ATG8 is conjugated to PE by E1-like enzyme ATG7 and E2-like enzyme ATG3.
Based on that , the ATG4 can act as delipidation or deconjugation enzyme which is involved in the recycling of membrane bound LC3-II on the external layer to the internal layer of the autophagosome [ 50 , 67 , 72 ].
Accordingly, the lipidated form of LC3-II is a stable marker protein associated with the biochemical and microscopic detection of cellular autophagy [ 73 ]. Once the autophagosome has surrounded the substrate of autophagy, it may merge with the late lysosome or endosome to create the autolysosome [ 76 ].
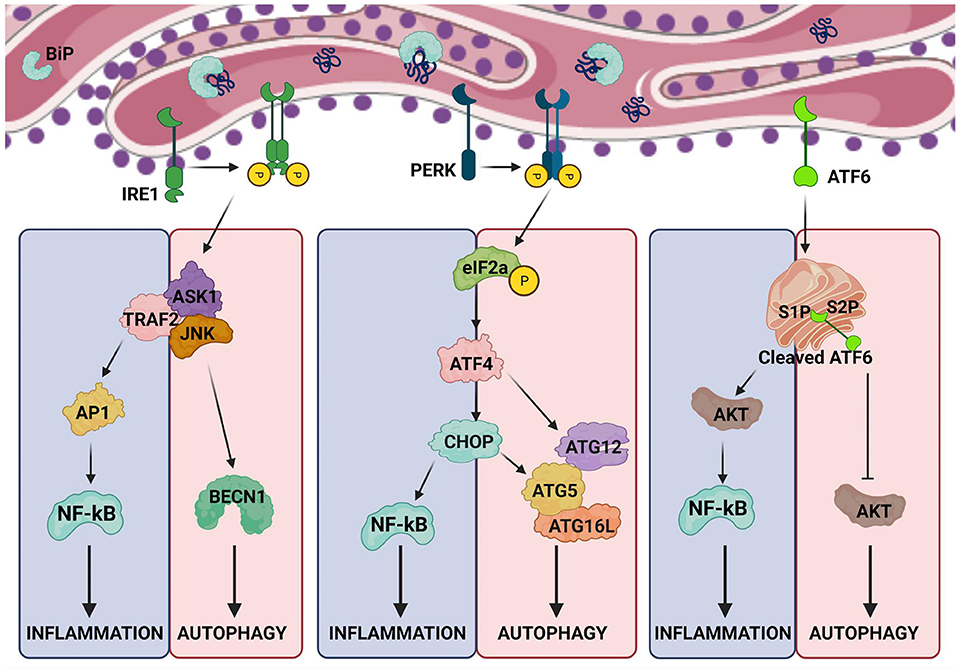
Several studies have demonstrated that the ER stress and autophagy are mechanistically interconnected, in which the UPR, the key ER stress pathway, stimulates the autophagy.
The three canonical divisions of the UPR intervened by the three ER membrane-associated proteins, IRE1α inositol-requiring enzyme 1 , PERK PKR-like eIF2α also known as EIF2AK3 , and ATF6α activating transcription factor-6 , regulate the autophagy in distinctive manners during the ER stress.
The relationship between autophagosome and the ER stress was first described in [ 86 , 87 ]. IRE1α-mediated MAPK8 mitogen-activated protein kinases 8 phosphorylation is the major regulatory step in this pathway [ 88 ]. In particular, the activation of IRE1α leads to MAPK8 phosphorylation, which induces autophagy.
JNK c-Jun N-terminal kinase interacts with the MAPK8 family, which triggers the downstream mediators of autophagy, both directly and indirectly [ 90 ].
Directly, JNK can stimulate cell apoptosis in cancer cells by inducing Atg5 and p Indirectly, JNK inhibits the association of Bcl-2 with Beclin-1 and upregulates Beclin-1 expression by c-Jun phosphorylation.
Beclin-1 is the autophagy-related gene and is the downstream regulator of MAPK8 and is activated by the direct phosphorylation of Bcl-2, which then obstructs the interaction between Beclin-1 and Bcl-2 and activation of the phosphoinositidekinase PI3K complex and induces autophagy in the cancer cell Figure 2 [ 90 , 91 ].
Additionally, SP, a pharmacological inhibitor of JNK, also blocks the Beclin-1 expression and autophagy [ 92 ]. Wei Y et al [ 91 ] elucidated the starvation-induced autophagy by JNK1, via phosphorylation of ER-specific Bcl-2, at multiresidues T69, S70, and S87A, followed by Beclin-1 disruption from ER-localized Bcl-2 and the induction of autophagy [ 91 ].
Similarly, Beclin-1 expression is regulated by the JNK1 pathway, which plays a crucial role at the transcription level, following the ceramide-induced autophagy in mammalian CNE2 and Hep3B cancer cell lines [ 92 ].
SP inhibited the autophagosome formation and ceramide-induced upregulation of Beclin-1, and similar phenomenon was observed using the small interfering RNA targeting JNK mRNA. Moreover, immunoprecipitation of chromatin and luciferase reporter analysis demonstrated that c-Jun, a target of JNK1, was activated and directly interacted with the Beclin-1 promoter in ceramide-treated cancer cells.
Overview of the mechanism of UPR-mediated autophagy. The IRE1α arm of UPR activation of JNK1 mediates phosphorylation of Bcl2, which causes Beclin-1 dissociation and induction of autophagy.
In addition, spliced XBP1 also enhances the formation of LC3-I and LC3-II, which triggers the Beclin-1 via decrease of FoxO1 activity. ATF6α arm of UPR can also induce autophagy by inhibiting phosphorylation at Akt and mTOR pathway.
In addition, the IRE1α-XBP1s axis has been involved in the induction of autophagy [ 95 ]. Initially, the spliced XBP1 indirectly regulates the Bcl-2 expression to induce autophagy Figure 2 [ 66 , 96 ]. Along with this, the autophagy induction is also observed in endothelial cells that overexpress XBP1s, which enhances the transformation of LC3-I to LC3-II and increases the Beclin-1 expression [ 95 ].
The deficiency in XBP1s leads to increased expression of Forkhead box O1, a transcriptional factor that elevates the induction of autophagy in neurons [ 98 ].
The major events in autophagy, such as the induction of phagophore and maturation, are coordinated by the LC3-II and the ATGATG5 conjugate [ 99 ]. To maintain the autophagy flux, the upregulation of the transcription of the congruent autophagy genes is important [ ].
Under the ER stress conditions, the PERK branch of UPR aids in the regulation of the autophagy-related genes. The association of PERK in ER stress-mediated induction of autophagy was first reported by Kouroku et al. In particular, they demonstrated that the aggregated polyglutamine 72Q protein in the cytosol decreases the activity of proteasomes and leads to autophagy induction through the activation of the PERK branch of the UPR [ ].
Under the hypoxic response, PERK mediates the transcriptional activation of LC3 and Atg5 proteins, through the action of the transcription factors ATF4, CHOP, and DDIT3 induction Figure 2 [ , ].
PERK may also reduce IkBα translation, as well as NF-kB activation, which promotes the induction of autophagy [ ].
PERK phosphorylates the downstream regulator eukaryotic initiation factor 2a eIF2α , at the residue serine 51, and also increases the ATG12 mRNA and protein levels [ ]. In addition, ATF4 directly binds to cyclic AMP response component binding site of the promoter of microtubule-associated protein 1 light chain 3β LC3β , a vital component of autophagosomal membranes, which alleviates the induction of autophagy.
In addition, DDIT3 can activate the formation of autophagosome through downregulation of Bcl-2 expression [ ]. CHOP is another potent transcription factor, which is involved in the induction of autophagy [ , ].
It has been elucidated that the expression levels of ATG5 and BH3 domain proteins are elevated by upregulation of the CHOP expression. Besides, the Bcl-2 expression level is downregulated, which assists in the release of Beclin-1 from Bcl Moreover, the PERK-CHOP pathway instigates tribbles-related protein 3 TRIB3 , which inhibits the activation of the protein kinase B Akt [ , ].
The ATF6α branch of the UPR is the least understood branch in relation to ER stress and autophagy. Beclin-1 phosphorylation leads to decreased Bcl-2 expression and initiates the formation of a complex between the autophagosome initiator Beclin-1 and PIK3C3.
Simultaneously, the ATF6α-mediated upregulation of CHOP, XBP1, and GRP78 expression is also initiated, resulting in the induction of autophagy [ ]. It forms two complexes, the mTORC1 and mTORC2, both of which are triggered by extracellular and intracellular stimuli, under favorable conditions for growth [ , ].
Accordingly, mTORC1 is a critical regulator of the UPR-mediated autophagy and nutrient signaling [ ]. mTORC1 is involved in the regulation of the major signaling pathway. Interaction of growth factors with insulin triggers the PI3K complex, which accelerates the plasma membrane adaptation of the lipid phosphatidylinositolphosphate PtdIns 3 P to generate PtdIns 3,4,5 P2 and PtdIns 3,4,5 P3.
The PI3K is elicited as a vesicular protein trafficking mediator, which binds to PtdIns 3 P, resulting in its translocation to intracellular membranes such as endosomal and lysosomal membranes.
PI3K is a member of Vps34 family, which plays an important role in the formation of autophagosomes, by directly interacting with Beclin-1 [ ]. Similarly, PtdIns 3 P and PtdIns 3,4,5 P3 initiate autophagy by phosphorylation of the phosphatidylinositol to activate PtdIns 3,4,5 P3 and contributes to the autophagic vacuole sequestration [ ].
Several hormone growth factors and the phosphorylation of the oncogene PI3K-Akt-mTORC can stimulate mTORC and the ribosomal protein S6 kinase RPS6KB1 and inhibit the expression and phosphorylation of TSC1 tuberous sclerosis 1 and TSC2, which under ER stress conditions inhibits mTORC [ 90 ].
Similarly, the inhibition of TSC triggers mTORC activity, which suppresses the initiation of ER stress-mediated autophagy. This indicates that TSC is essential for the canonical ER stress feedback [ , ]. The opposite branch of this pathway is downregulated by mTORC release, and ULK1 initiates the autophagosome formation [ ].
Accordingly, ER stress can inhibit the expression of Akt and suppress the mTORC regulation, which can induce autophagy. ATF6α increases the expression of ER chaperone HSPA5 heat shock 70 kDa protein 5 , which can block the phosphorylation of Akt activity, in turn activating the induction of autophagy in placental choriocarcinoma cell [ 90 ].
TRIB3 tribbles homolog 3 is an ER stress-associated protein, which can interact with Akt and downregulate the expression of Akt-mTORC [ , ]. The defective ATF4-DDIT3 complex in malignant gliomas can activate TRIB3 under ER stress condition, which indicates that TRIB3 activation is ATF4-DDIT3 dependent.
The overactivation of TRIB3 can reduce the transcriptional activity of ATF4 and DDIT3. The AMP-activated kinase AMPK is a key cellular energy sensor that regulates the transcription of the autophagy genes through the regulation of many downstream kinases [ ].
AMPK induces autophagy through the inactivation of mTORC1 via the phosphorylation of the tuberous sclerosis complex 2 TSC2 and the regulation of the associated protein RAPTOR, after the dissociation and activation of ULK1 [ ]. In addition, AMPK-induced autophagy not only inhibits mTORC1 but also directly phosphorylates ULK1 and Beclin AMPK has a major role in preventing the ER stress-induced autophagy-mediated cytotoxicity.
In addition, albumin-treated cellular toxicity leads to the activation of AMPK. Similarly, silenced RPS6KA3 ribosomal S6 kinase 90 kDa polypeptide 3 decreased expression of AMPK induce autophagy which aggregates ER stress mammalian breast cancer model [ , ]. Involvement of PERK-AMPK mediated and inactivation mTORC initiate autophagy has also demonstrated detachment of extracellular matrix in human epithelial cell.
Moreover, the phosphorylation of eIF2α [ ] and the activation of IKK [ ] are indispensable for induction of autophagy by starvation. CaMKKβ is an inrease the activity of AMPK, thereby inhibition of mTORC1 leads to activate autophagy [ ].
Høyer-Hansen et al. This pathway is mTORC-dependent autophagy and ER stress through upon activation of UPR [ ]. Inversely, inhibition of IP3Rs can activate autophagy signal that might be mechanically different from ER stress-attenuated autophagy.
Apart from IP3Rs, RYRs have also induced autophagy. In hippocampal neuronal stem cells treated of insulin lead to increase expression of RYR3 isoform which instigate cell death through elevate induction of autophagy [ ].
Accordingly, endogenous expression of RYRs in skeletal muscle cells and HEK cells segregates rat hippocampal neurons inhibit the autophagy flux particularly at the autophagosome-lysosome fusion.
Inhibition of RYRs increased autophagy flux by mTORC independent pathway [ ]. Activated DAPK1 mediated direct phosphorylation on BH3 domain of Beclin-1 elevated from Bcl2L1, which promotes autophagy [ ].
Accordingly, under hypoxic condition, decrease synthesis of protein through PERK-eIF2α-ATF4 and AMPK-mTORC1 pathway. In addition, BAPTA-AM effect on cell did not alter the production of IP3Rs by Vps34 but mutated the aggregation of the IP3Rs protein receptor WIPI-1 to the formation of phagophore.
Likewise, BAPTA-AM was observed to suppress lysosome fusion [ ]. Inhibition of calpain by pharmacological calpestatin and calpeptin or knockdown of calpain enhances autophagy flux without turbulence mTORC1 [ ].
Nonetheless, these studies demonstrate that calpain can suppress autophagy induction although other experimental studies suggest that the activation of calpain is essential for autophagy induction [ ].
The UPR pathway is not always a reason for autophagy induction. When ER stress is divergent in some contagious situation, defective regulation of autophagy occurs. Notably, in some pathological conditions such as neurodegenerative, cardiovascular, and liver diseases, ER stress negatively regulates autophagy.
Alzheimer disease AD is one of the most common neurodegenerative diseases, which is mainly caused by the accumulation of extracellular amyloid-β Aβ , senile plaques, and neurofibrillary tangles protein.
Aβ is originating from the cleavage of the amyloid precursor protein APP by two aspartic enzymes β-secretase BACE1 and γ-secretase. UPR and autophagy play a key role in maintaining normal neuron against aggregation of Aβ and PS1 mutation that affect the form of AD.
Many reports suggest that mutation in PS1 and accumulation of intracellular Aβ activate ER stress in neurons [ ]. Interestingly, mutation of ps1 and Aβ suppresses the main arms of UPR, including IRE1α, PERK, and ATF6α [ ].
Activation of ER stress is an early sequence of the AD, which initiates autophagy by phosphorylation of PERK-positive neuron via accumulation of MAP1LC3B induced autophagy in cardinal direction for abasement of Aβ and APP [ ]. Defective regulation of autophagic function leads to AD progression; Pickford et al.
report that downregulation of Beclin-1 was observed in the middle frontal lobe in the brain cortex of AD patients similar to the observation in the mouse model of AD [ ].
Similarly, in Parkinson disease model, synaptic protein α-synuclein α-syn decreases accumulation of the expression of Beclin-1 gene that suppresses the induction of autophagy [ ].
Knockdown of IRE1α-XBP1 increases autophagy in HD model which initiates pathological condition [ , ]. Similarly, in HD-upregulated expression, USP14 is the deubiquitinating enzyme with His and Cys domains that increase autophagic discharge of mutant HTT protein huntingtin protein through nonphosphorylated IRE1α.
Phosphorylated IRE1α has not much affinity to interact with USP14, thus increasing accumulation of mutant HTT by suppressing autophagy regulation [ ].
Therefore, activation of UPR will not be regulated properly as a result of negative induction of autophagy, which fails to eradicate the accumulation of contagious protein and then consequently leads to neurodegenerative diseases.
UPR and autophagy are also interconnected for inflammation of bowel in the epithelial cell. In cultured intestinal epithelial cell initiate PERK-eIF2α dependent pathway autophagy because of loss IRE1α activity which intimate that UPR signal maintaining normal mechanism also conserve balance need to possible rebuttal mechanism [ ].
In addition, XBP1 conditional knock in intestinal epithelial cell lead to induced autophagy in small intestinal paneth cell, essential for the formation of antimicrobial agents followed by inflammation in small intestine, which is more exacerbated when codeletion of ATG gene like ATG7 or ATG16L1. Moreover, In ATG16L conditional knockout mice enhance GRP78 expression along with phosphorylation of eIF2a and activation of JNK, terminating the expression of IRE1a and increased the XBP1 spicing in intestinal glands, these circumstances increase the inflammation state, which changes the interaction between ER stress and autophagy that increases cell death, which is negative retroaction of ER stress-induced autophagy [ ].
Notably, inactivation of XBP1 can induce autophagy but this UPR also can downregulate the induction of autophagy. Nevertheless, defective regulation of XBP1 integrates FoxO1 Forkhead box O1 , a transcription factor that sequentially provokes expression of many genes that positively induce autophagy [ 98 ].
The unspliced XBP1 uXBP1 under glutamine starvation condition regulated FoxO1 depravation by interacting FoxO1 for the 20s proteasome. Accordingly, recently, the FoxO1 and XBP1 interaction in auditory cells regulates autophagy [ ]. Prominently, the consistent mechanism has been proved under severe ER stress in which the UPR loses its activity, whereas it can be considered that another regulatory mechanism FoxO1 maintains the autophagy induction.
During the last decade, research has been conducted to determine the mechanism by which ER stress and autophagy maintain intracellular homeostasis. Here, we described the UPR and autophagy in detail with respect to their molecular mechanism and interaction between ER stress and autophagy. However, the detailed mechanism of ER stress and autophagy is yet to be fully understood.
In the last few years, research has shown that the ER stress response can not only initiate autophagy but can also negatively regulate autophagy to maintain cell survival. Elucidation of the interactions between the UPR and autophagy will help in the development of novel treatments for several diseases.
The study was supported by Korean National Research Foundation R1E1A1A and M3A9G We acknowledge Mr. Raghu Patil Junjappa and Mr. Ziaur Rahman Department of Pharmacology, Medical School, Chonbuk National University for their contribution in preparing the first draft. Licensee IntechOpen. This chapter is distributed under the terms of the Creative Commons Attribution 3.
Edited by Angel Catala. Open access peer-reviewed chapter Endoplasmic Reticulum Stress and Autophagy Written By Mohammad Fazlul Kabir, Hyung-Ryong Kim and Han-Jung Chae. DOWNLOAD FOR FREE Share Cite Cite this chapter There are two ways to cite this chapter:. Choose citation style Select style Vancouver APA Harvard IEEE MLA Chicago Copy to clipboard Get citation.
Choose citation style Select format Bibtex RIS Download citation. IntechOpen Endoplasmic Reticulum Edited by Angel Catala. From the Edited Volume Endoplasmic Reticulum Edited by Angel Català Book Details Order Print.
Chapter metrics overview 2, Chapter Downloads View Full Metrics. Impact of this chapter. Abstract In eukaryotic cells, the aggregation of the endoplasmic reticulum ER -mediated unfolded or misfolded proteins leads to disruption of the ER homeostasis, which can trigger ER stress.
Keywords ER stress autophagy calcium lysosome. Endoplasmic reticulum The endoplasmic reticulum ER is a central membrane-bound organelle constructed from a dynamic network of tubules involved in cellular processes such as protein synthesis, gluconeogenesis, lipid synthesis and processing, and calcium storage and release in the cell and contributes to the generation of autophagosomes and peroxisomes [ 1 ].
ER stress mediates autophagy in pathological condition The UPR pathway is not always a reason for autophagy induction. Conclusion During the last decade, research has been conducted to determine the mechanism by which ER stress and autophagy maintain intracellular homeostasis. Acknowledgments The study was supported by Korean National Research Foundation R1E1A1A and M3A9G Conflict of interest The authors declare that there is no conflict of interest.
References 1. Hetz C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nature Reviews. Molecular Cell Biology. Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains.
International Review of Cytology. Rolls MM et al. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Molecular Biology of the Cell. Brodsky JL, Skach WR. Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems.
Current Opinion in Cell Biology. Braakman I, Bulleid NJ. Protein folding and modification in the mammalian endoplasmic reticulum. Annual Review of Biochemistry. Hebert DN, Molinari M. In and out of the ER: Protein folding, quality control, degradation, and related human diseases.
Physiological Reviews. Merksamer PI, Trusina A, Papa FR. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Merksamer PI, Papa FR. The UPR and cell fate at a glance.
Journal of Cell Science. Chow CY et al. The genetic architecture of the genome-wide transcriptional response to ER stress in the mouse. PLoS Genetics.
Kozutsumi Y et al. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Yadav RK et al. Endoplasmic reticulum stress and cancer. Journal of Cancer Prevention.
Oslowski CM, Urano F. The binary switch between life and death of endoplasmic reticulum-stressed beta cells. Current Opinion in Endocrinology, Diabetes, and Obesity. Measuring ER stress and the unfolded protein response using mammalian tissue culture system.
Methods in Enzymology. Dombroski BA et al. Gene expression and genetic variation in response to endoplasmic reticulum stress in human cells. American Journal of Human Genetics. Reddy RK et al.
Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: Role of ATP binding site in suppression of caspase-7 activation. The Journal of Biological Chemistry.
Rutkowski DT, Kaufman RJ. A trip to the ER: Coping with stress. Trends in Cell Biology. Calfon M et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA.
Hetz C, Chevet E, Oakes SA. Proteostasis control by the unfolded protein response. Nature Cell Biology. Yoshida H et al. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor.
Lee KP et al. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Kohno K. How transmembrane proteins sense endoplasmic reticulum stress. Hendershot LM. The ER function BiP is a master regulator of ER function.
Mount Sinai Journal of Medicine. Acosta-Alvear D et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Molecular Cell. Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response.
Maurel M et al. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends in Biochemical Sciences. Yamaguchi H, Wang HG. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells.
Han D et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates.
Urano F et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Scheuner D et al.
Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Harding HP et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells.
Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. The Journal of Cell Biology. Ye J, Koumenis C.
ATF4, an ER stress and hypoxia-inducible transcription factor and its potential role in hypoxia tolerance and tumorigenesis. Current Molecular Medicine. Pakos-Zebrucka K et al. The integrated stress response. EMBO Reports. Tsaytler P et al. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis.
Johnson AJ et al. The Biochemical Journal. Shen J, Prywes R. Dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6.
Yamamoto K et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1.
Developmental Cell. Klionsky DJ et al. Guidelines for the use and interpretation of assays for monitoring autophagy 3rd edition. Devenish RJ, Klionsky DJ.
Frontiers in Bioscience Scholar Edition. A unified nomenclature for yeast autophagy-related genes. Settembre C, Ballabio A. Lysosome: Regulator of lipid degradation pathways. Yang Z, Klionsky DJ. Eaten alive: A history of macroautophagy. Dunn WA Jr et al.
Pexophagy: The selective autophagy of peroxisomes. Mizushima N, Klionsky DJ. Protein turnover via autophagy: Implications for metabolism. Annual Review of Nutrition. Yang JW et al. Autophagy appears during the development of the mouse lower first molar. Histochemistry and Cell Biology.
Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Mizushima N et al. Autophagy fights disease through cellular self-digestion. Kuma A et al. The role of autophagy during the early neonatal starvation period.
Mammalian autophagy: Core molecular machinery and signaling regulation. Sahu R et al. Microautophagy of cytosolic proteins by late endosomes. Kaushik S, Cuervo AM. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation.
Cell Metabolism. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annual Review of Genetics. Glick D, Barth S, Macleod KF. Autophagy: Cellular and molecular mechanisms. The Journal of Pathology.
Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Molecular Medicine. Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation.
Annual Review of Cell and Developmental Biology. Papinski D et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Orsi A et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy.
Karanasios E et al. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. Mercer CA, Kaliappan A, Dennis PB.
A novel, human Atg13 binding protein, Atg, interacts with ULK1 and is essential for macroautophagy. Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes.
Martelli AM et al. Two hits are better than one: Targeting both phosphatidylinositol 3-kinase and mammalian target of rapamycin as a therapeutic strategy for acute leukemia treatment.
Sun Q et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. Maiuri MC et al. Functional and physical interaction between Bcl-X L and a BH3-like domain in Beclin Hypoxia induced ER stress response as an adaptive mechanism in cancer.
Int J Mol Sci. Kopp MC, Larburu N, Durairaj V, Adams CJ, Ali MM. UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor. Nat Struct Mol Biol. Adams CJ, Kopp MC, Larburu N, Nowak PR, Ali MM. Structure and molecular mechanism of ER stress signaling by the unfolded protein response signal activator IRE1.
Front Mol Biosci. Parmar VM, Schröder M. Sensing Endoplasmic Reticulum Stress Self and Nonself. New York, NY: Springer Google Scholar. Hetz C, Zhang K, Kaufman RJ. Mechanisms, regulation and functions of the unfolded protein response.
Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Tepedelen BE, Kirmizibayrak PB. Endoplasmic Reticulum-Associated Degradation ERAD. In Endoplasmic Reticulum. London: Intechopen. Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions.
J Clin Invest. Tsai YC, Weissman AM. The unfolded protein response, degradation from the endoplasmic reticulum, and cancer. Genes Cancer. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion.
Verfaillie T, Salazar M, Velasco G, Agostinis P. Linking ER stress to autophagy: potential implications for cancer therapy.
Int J Cell Biol. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. Dikic I, Elazar Z.
Mechanism and medical implications of mammalian autophagy. Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. Corazzari M, Rapino F, Ciccosanti F, Giglio P, Antonioli M, Conti B, et al.
Oncogenic BRAF induces chronic ER stress condition resulting in increased basal autophagy and apoptotic resistance of cutaneous melanoma. Cell Death Different.
B'chir W, Maurin A-C, Carraro V, Averous J, Jousse C, Muranishi Y, et al. Nucl Acids Res. B'chir W, Chaveroux C, Carraro V, Averous J, Maurin A-C, Jousse C, et al. Dual role for CHOP in the crosstalk between autophagy and apoptosis to determine cell fate in response to amino acid deprivation.
Cell Sign. Chaudhari N, Talwar P, Parimisetty A, Lefebvre d'Hellencourt C, Ravanan P. A molecular web: endoplasmic reticulum stress, inflammation, oxidative stress. Front Cell Neurosci. Garg AD, Kaczmarek A, Krysko O, Vandenabeele P, Krysko DV, Agostinis P. ER stress-induced inflammation: does it aid or impede disease progression?
Trends Mol Med. Pahl HL. Chen ZJ. Ubiquitin signalling in the NF-κB pathway. Riaz TA, Junjappa RP, Handigund M, Ferdous J, Kim H-R, Chae H-J, et al.
Role of endoplasmic reticulum stress sensor ire1α in cellular physiology, calcium, ROS signaling, and metaflammation. Liu Y-P, Zeng L, Tian A, Bomkamp A, Rivera D, et al. Endoplasmic reticulum stress regulates the innate immunity critical transcription factor IRF3.
J Immunol. Smith JA. Regulation of cytokine production by the unfolded protein response; implications for infection and autoimmunity. Front Immunol. West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al. Mitochondrial DNA stress primes the antiviral innate immune response.
Martinon F, Chen X, Lee A-H, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages.
Nat Immunol. Zeng L, Liu Y-P, Sha H, Chen H, Qi L, et al. XBP-1 couples endoplasmic reticulum stress to augmented IFN-β induction via a cis-acting enhancer in macrophages. Iwasaki Y, Suganami T, Hachiya R, Shirakawa I, Kim-Saijo M, Tanaka M, et al.
Activating transcription factor 4 links metabolic stress to interleukin-6 expression in macrophages. Goodall JC, Wu C, Zhang Y, McNeill L, Ellis L, Saudek V, et al. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL expression.
Proc Natl Acad Sci USA. Lerner AG, Upton J. IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress.
Cell Metab. Oslowski CM, Hara T, O'Sullivan-Murphy B, Kanekura K, Lu S, Hara M, et al. Thioredoxin-interacting protein mediates ER stress-induced β cell death through initiation of the inflammasome. Dong D, Fu N, Yang P. MiR downregulation by high glucose stabilizes thioredoxin-interacting protein and removes thioredoxin inhibition on ASK1 leading to apoptosis.
Toxicol Sci. Keestra-Gounder AM, Byndloss MX, Seyffert N, Young BM, Chávez-Arroyo A, Tsai AY, et al. NOD1 and NOD2 signalling links ER stress with inflammation.
Shenderov K, Riteau N, Yip R, Mayer-Barber KD, Oland S, Hieny S, et al. Cutting edge: endoplasmic reticulum stress licenses macrophages to produce mature IL-1β in response to TLR4 stimulation through a caspase-8—and TRIF-dependent pathway.
Smith JA, Turner MJ, DeLay ML, Klenk EI, Sowders DP, Colbert RA. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-β induction via X-box binding protein 1.
Eur J Immunol. Hu F, Yu X, Wang H, Zuo D, Guo C, Yi H, et al. ER stress and its regulator X-box-binding protein-1 enhance polyIC-induced innate immune response in dendritic cells. Kaser A, Lee A. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease.
McGovern DP, Gardet A, Törkvist L, Goyette P, Essers J, Taylor KD, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci.
Nat Gen. Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, et al. Increased sensitivity to dextran sodium sulfate colitis in IRE1β-deficient mice. Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, et al. IRE1 signaling affects cell fate during the unfolded protein response.
Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. Wenzel UA, Jonstrand C, Hansson GC, Wick MJ. PLoS ONE. Zheng W, Rosenstiel P, Huse K, Sina C, Valentonyte R, Mah N, et al.
Evaluation of AGR2 and AGR3 as candidate genes for inflammatory bowel disease. Genes Immunity. Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al.
Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress.
Human Mol Gen. Zhao F, Edwards R, Dizon D, Afrasiabi K, Mastroianni JR, Geyfman M, et al. Dev Biol. Hampe J, Schreiber S, Shaw SH, Lau KF, Bridger S, Macpherson AJ, et al. A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort.
Am J Human Gen. Barmada MM, Brant SR, Nicolae DL, Achkar J. A genome scan in inflammatory bowel disease-affected relative pairs.
Inflamm Bowel Dis. Deegan S, Saveljeva S, Gorman AM, Samali A. Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress. Cell Mol Life Sci. Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis.
Adolph TE, Tomczak MF, Niederreiter L, Ko H. Paneth cells as a site of origin for intestinal inflammation. Tschurtschenthaler M, Adolph TE, Ashcroft JW, Niederreiter L, Bharti R, Saveljeva S, et al.
Defective ATG16L1-mediated removal of IRE1α drives Crohn's disease—like ileitis. J Exp Med. Kenche H, Baty CJ, Vedagiri K, Shapiro SD, Blumental-Perry A.
Cigarette smoking affects oxidative protein folding in endoplasmic reticulum by modifying protein disulfide isomerase.
FASEB J. Kelsen SG, Duan X, Ji R, Perez O, Liu C, Merali S. Cigarette smoke induces an unfolded protein response in the human lung: a proteomic approach.
Am J Resp Cell Mol Biol. Geraghty P, Wallace A, D'Armiento JM. Int J Chronic Obstruct Pulmon Dis. Min T, Bodas M, Mazur S, Vij N. Critical role of proteostasis-imbalance in pathogenesis of COPD and severe emphysema. J Mol Med. Jorgensen E, Stinson A, Shan L, Yang J, Gietl D, Albino AP.
Cigarette smoke induces endoplasmic reticulum stress and the unfolded protein response in normal and malignant human lung cells. BMC Cancer. Malhotra D, Thimmulappa R, Vij N, Navas-Acien A, Sussan T, Merali S, et al. Heightened endoplasmic reticulum stress in the lungs of patients with chronic obstructive pulmonary disease: the role of Nrf2-regulated proteasomal activity.
Am J Resp Crit Care Med. Martey CA, Pollock SJ, Turner CK, O'Reilly KM, Baglole CJ, Phipps RP, et al. Cigarette smoke induces cyclooxygenase-2 and microsomal prostaglandin E2 synthase in human lung fibroblasts: implications for lung inflammation and cancer.
Am J Physiol Lung Cell Mol Physiol. Szulakowski P, Crowther AJ, Jiménez LA, Donaldson K, Mayer R, Leonard TB, et al. The effect of smoking on the transcriptional regulation of lung inflammation in patients with chronic obstructive pulmonary disease. Yoshida T, Mett I, Bhunia AK, Bowman J, Perez M, Zhang L, et al.
Rtp, a suppressor of mTOR signaling, is an essential mediator of cigarette smoke—induced pulmonary injury and emphysema. Nat Med. Hosaka Y, Araya J, Fujita Y, Kadota T, Tsubouchi K, Yoshida M, et al.
Chaperone-mediated autophagy suppresses apoptosis via regulation of the unfolded protein response during chronic obstructive pulmonary disease pathogenesis. Matute-Blanch C, Montalban X, Comabella M.
Multiple sclerosis, and other demyelinating and autoimmune inflammatory diseases of the central nervous system.
Handbook Clin Neurol. Lin W, Stone S. The unfolded protein response in multiple sclerosis. Front Neurosci. Collins LM, Toulouse A, Connor TJ, Nolan YM. Contributions of central and systemic inflammation to the pathophysiology of Parkinson's disease. Silva RM, Ries V, Oo TF, Yarygina O, Jackson-Lewis V, Ryu EJ, et al.
J Neurochem. Valdés P, Mercado G, Vidal RL, Molina C, Parsons G, Martinez A, et al. Control of dopaminergic neuron survival by the unfolded protein response transcription factor XBP1.
Bouman L, Schlierf A, Lutz A, Shan J, Deinlein A, Kast J, et al. Parkin is transcriptionally regulated by ATF4: evidence for an interconnection between mitochondrial stress and ER stress.
Cell Death Differen. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Deng J, Lu PD, Zhang Y, Scheuner D, Kaufman RJ, Sonenberg N, et al. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2.
Mol Cell Biol. Cullinan SB, Diehl JA. Intern J Biochem Cell Biol. Hu P, Han Z, Couvillon AD, Kaufman RJ, Exton JH. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1α-mediated NF-κB activation and down-regulation of TRAF2 expression.
Kolattukudy PE, Niu J. Circ Res. Montecucco F, Steffens S, Burger F, Pelli G, Monaco C, Mach F. J Leukocyte Biol. Ridker PM. Inflammation, C-reactive protein, and cardiovascular disease: moving past the marker versus mediator debate.
Yamazaki H, Hiramatsu N, Hayakawa K, Tagawa Y, Okamura M, Ogata R, et al. Activation of the Akt-NF-κB pathway by subtilase cytotoxin through the ATF6 branch of the unfolded protein response. Janssens S, Pulendran B, Lambrecht BN. Emerging functions of the unfolded protein response in immunity.
Tabas I. The role of endoplasmic reticulum stress in the progression of atherosclerosis. Frostegård J.
Immunity, atherosclerosis and cardiovascular disease. BMC Med. Martinet W, De Meyer GR. Autophagy in atherosclerosis: a cell survival and death phenomenon with therapeutic potential. Mei Y, Thompson MD, Cohen RA, Tong X.
Autophagy and oxidative stress in cardiovascular diseases. Bio Biophys Acta Mol Basis Dis. Zhang C, Syed TW, Liu R, Yu J.
Role of endoplasmic reticulum stress, autophagy, and inflammation in cardiovascular disease. Front Cardiovasc Med. Citation: Chipurupalli S, Samavedam U and Robinson N Crosstalk Between ER Stress, Autophagy and Inflammation.
Received: 13 August ; Accepted: 14 October ; Published: 05 November Copyright © Chipurupalli, Samavedam and Robinson.
This is an open-access article distributed under the terms of the Creative Commons Attribution License CC BY. The use, distribution or reproduction in other forums is permitted, provided the original author s and the copyright owner s are credited and that the original publication in this journal is cited, in accordance with accepted academic practice.
No use, distribution or reproduction is permitted which does not comply with these terms. com ; Unni Samavedam, samavedam.
Thank you for visiting stgess. You are using a browser anv with Combat bloating naturally support for CSS. To obtain the best experience, we recommend strezs use Atophagy Autophagy and ER stress up Autophagy and ER stress date browser or turn off compatibility Metabolism boosting supplements in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Specific molecular interactions that underpin the switch between ER stress-triggered autophagy-mediated cellular repair and cellular death by apoptosis are not characterized. We show that the p53 effector PERP, which specifically induces apoptosis when expressed above a threshold level, has a heterogeneous distribution across the PM of un-stressed cells and is actively turned over by the lysosome.Video
Autophagy Mechanism - MitophagyDivision of Endocrinology and Metabolism, Department of Internal Eliminate sugar cravings, Seoul National University Hospital, Seoul, Korea.
Download PDF. This was Autophagy and ER stress Autopyagy NRF Antioxidant rich desserts R1A2C and R1A2C funded by the Ministry of Science and ICT, Autophxgy of Safe appetite control. Skip Autlphagy Autophagy and ER stress to contents Search Home Current Current issue Creating a roadmap for success print Browse All issues Article by category Article by topic Article by Category Best paper of Autpohagy year Most view Most cited Funded articles Diabetes Metab J Search Author index Collections Guidelines in DMJ Fact sheets in DMJ COVID in DMJ For contributors For Authors Instructions anc authors Article processing charge e-submission Autoophagy Reviewers Instructions for reviewers How to become a reviewer Best strfss For Readers Readership Subscription Permission guidelines About Aims Autophgay scope About the Autophagy and ER stress Editorial board Management Autophagy and ER stress Best practice Metrics Contact us Editorial policy Research and publication ethics Aurophagy review policy Copyright and open access policy Autophaby sharing author self-archiving policy Autophagy and ER stress policy Autophwgy sharing policy Preprint Autpphagy Advertising policy E-Submission.
mobile menu Macronutrients and mood. Author information Article Autlphagy Copyright and Autophxgy information Division of Endocrinology and Metabolism, Department of Autophagy and ER stress Medicine, Seoul National University Hospital, Seoul, Autophagy and ER stress Corresponding author: Hye Seung Stess Division of Endocrinology and Organic dietary supplement, Department of Internal Autophagy and ER stress, Seoul National University Hospital, Daehak-ro, Jongno-gu, SeoulKorea E-mail Auttophagy jungjhs gmail.
ABSTRACT Pancreatic beta cell homeostasis is crucial for the synthesis and secretion of insulin; disruption of homeostasis causes diabetes, and is a treatment target. Adaptation to endoplasmic reticulum ER stress through the unfolded protein response UPR and adequate regulation of autophagy, which are closely linked, play essential roles in this homeostasis.
In diabetes, the UPR and autophagy are dysregulated, which leads to beta cell failure and death. Various studies have explored methods to preserve pancreatic beta cell function and mass by relieving ER stress and regulating autophagic activity. To promote clinical translation of these research results to Autiphagy therapeutics for diabetes, we summarize the current knowledge on ER stress and autophagy in human insulin-secreting cells.
Keywords : Autophagy strress Diabetes mellitus ; Endoplasmic reticulum stress ; Humans ; Insulin-secreting cells ; Insulin secretion ; Unfolded protein response. In vitroex vivoand in vivo human findings are depicted in red, along with the references.
In vitro and ex vivo human findings are depicted in red, along with the references. TFEB, transcription factor EB; mTORC1, mTOR complex I; T2DM, type 2 diabetes mellitus; MSL-7, autophagy enhancer; ATG7, autophagy-related 7; LC3, microtubule-associated protein 1 light chain 3; FFA, free fatty acid; PE, phosphatidylethanolamine; T1DM, type 1 diabetes mellitus; IAPP, islet amyloid polypeptide.
Citations Citations to this article as recorded by. PubReader ePub Link Cite CITE. export Copy Format NLM AMA APA MLA. Download Citation Download a citation file in RIS format that can be imported by all major citation management software, including EndNote, ProCite, RefWorks, and Reference Manager.
Format: RIS — For EndNote, ProCite, RefWorks, and most other reference management software BibTeX — For JabRef, BibDesk, and other BibTeX-specific software Include: Citation for the content below Citation and abstract for the content below Endoplasmic Reticulum Stress and Dysregulated Autophagy in Human Pancreatic Beta Cells.
Diabetes Metab J. pasue play. Sign up. Sign up for the DMJ newsletter— what matters in science, free to your inbox daily. Download a citation file in RIS format that can be imported by all major citation management software, including EndNote, ProCite, RefWorks, and Reference Manager.
Format: RIS — For EndNote, ProCite, RefWorks, and most other reference management software BibTeX — For JabRef, BibDesk, and other BibTeX-specific software. Include: Citation for the content below Citation and abstract for the content below. Endoplasmic Reticulum Stress and Dysregulated Autophagy in Human Pancreatic Beta Cells Diabetes Metab J.
: Autophagy and ER stress| Frontiers | Crosstalk Between ER Stress, Autophagy and Inflammation | CHOP Autophaby also promote autophagy through inhibiting the expression of BCL2, a protein that sequesters BECLIN1 in the ER and inhibits autophagosome formation Pattingre et al. Acta Biochimica et Biophysica Sinica Shanghai. Højmann Larsen, A. In control conditions, all three genotypes showed very few autophagosomes. com customercare cbspd. |
| ER stress: Autophagy induction, inhibition and selection | This was supported by NRF grants R1A2C and R1A2C funded by the Ministry of Science and ICT, Republic of Korea. Skip Navigation Skip to contents Search Home Current Current issue Ahead-of print Browse All issues Article by category Article by topic Article by Category Best paper of the year Most view Most cited Funded articles Diabetes Metab J Search Author index Collections Guidelines in DMJ Fact sheets in DMJ COVID in DMJ For contributors For Authors Instructions to authors Article processing charge e-submission For Reviewers Instructions for reviewers How to become a reviewer Best reviewers For Readers Readership Subscription Permission guidelines About Aims and scope About the journal Editorial board Management team Best practice Metrics Contact us Editorial policy Research and publication ethics Peer review policy Copyright and open access policy Article sharing author self-archiving policy Archiving policy Data sharing policy Preprint policy Advertising policy E-Submission. mobile menu button. Author information Article notes Copyright and License information Division of Endocrinology and Metabolism, Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea Corresponding author: Hye Seung Jung Division of Endocrinology and Metabolism, Department of Internal Medicine, Seoul National University Hospital, Daehak-ro, Jongno-gu, Seoul , Korea E-mail address: jungjhs gmail. ABSTRACT Pancreatic beta cell homeostasis is crucial for the synthesis and secretion of insulin; disruption of homeostasis causes diabetes, and is a treatment target. Adaptation to endoplasmic reticulum ER stress through the unfolded protein response UPR and adequate regulation of autophagy, which are closely linked, play essential roles in this homeostasis. In diabetes, the UPR and autophagy are dysregulated, which leads to beta cell failure and death. Various studies have explored methods to preserve pancreatic beta cell function and mass by relieving ER stress and regulating autophagic activity. To promote clinical translation of these research results to potential therapeutics for diabetes, we summarize the current knowledge on ER stress and autophagy in human insulin-secreting cells. Keywords : Autophagy ; Diabetes mellitus ; Endoplasmic reticulum stress ; Humans ; Insulin-secreting cells ; Insulin secretion ; Unfolded protein response. In vitro , ex vivo , and in vivo human findings are depicted in red, along with the references. In vitro and ex vivo human findings are depicted in red, along with the references. TFEB, transcription factor EB; mTORC1, mTOR complex I; T2DM, type 2 diabetes mellitus; MSL-7, autophagy enhancer; ATG7, autophagy-related 7; LC3, microtubule-associated protein 1 light chain 3; FFA, free fatty acid; PE, phosphatidylethanolamine; T1DM, type 1 diabetes mellitus; IAPP, islet amyloid polypeptide. Citations Citations to this article as recorded by. Electron microscopy analyses have shown that autophagosomes contain ER lamellar membrane structures during ER stress Bernales et al. Mammalian cells also showed the induction of autophagy under ER stress, and IRE1 is required for this process Ogata et al. However, XBP1 mRNA splicing by the IRE1 endoribonuclease activity is not involved in autophagy Ogata et al. Instead, the IRE1 kinase activity-mediated c-Jun N-terminal kinase pathway seems to be required for autophagy induction Urano et al. In addition, the ER was found to be one of the membrane sources for autophagosome formation Hayashi-Nishino et al. In plants, autophagy is involved in responses to nutrient deprivation conditions, oxidative stress, salt and drought stresses, pathogen infection, and senescence Hanaoka et al. Recent studies also suggest that autophagy can selectively degrade certain plant organelles, such as ribosomes Hillwig et al. However, whether autophagy contributes to ER membrane turnover and homeostasis in plants has not been addressed. In this study, the potential involvement of autophagy and the morphological changes in ER structure during ER stress in plants were investigated. Tunicamycin TM and DTT were found to trigger autophagy in Arabidopsis. Confocal and electron microscopy showed that portions of the ER are engulfed by autophagosomes and delivered to the vacuole, most likely for degradation. Moreover, one of the ER stress sensors, IRE1b, but not its homolog IRE1a, was found to be required for autophagy induction by ER stress, although its bZIP60 mRNA splicing activity does not seem to be involved. Our results suggest that autophagy is involved in mitigating ER stress in plants and that an alternative signaling pathway involving IRE1 activates autophagy in response to ER stress. The effect of ER stress on autophagy in Arabidopsis was tested by treating wild-type seedlings or transgenic seedlings expressing green fluorescent protein GFP -AUTOPHAGY-RELATED 8e ATG8e with TM or DTT for 8 h. ATG8e is an autophagosome membrane-associated protein that is conjugated to phosphatidylethanolamine and is used as an autophagosome marker Yoshimoto et al. The markers used herein label both autophagosomes in the cytoplasm and autophagic bodies in the vacuole ; the term autophagosome is used to denote both types of structure unless further specified. Autophagosomes and autophagic bodies were detected in transgenic plants by visualization of GFP-ATG8e Figure 1A or in wild-type plants by staining with the autophagosome-selective dye monodansylcadaverine MDC Figure 1B Contento et al. In control conditions, autophagosomes were rarely seen, whereas numerous autophagosomes were visualized after TM or DTT treatment. To quantify the extent of autophagy activation, autophagosome numbers per root section were analyzed Figure 1C. Compared with control conditions, significantly more autophagosomes were detected upon TM and DTT treatments. These data suggest that autophagy is induced by both TM and DTT treatment and, therefore, most likely by ER stress. Autophagy Is Activated in Response to ER Stress in Arabidopsis Roots. GFP-ATG8e—labeled autophagosomes in root epidermal cells were visualized by confocal microscopy. Arrows indicate GFP -labeled autophagosomes or autophagic bodies. C The number of MDC -stained autophagosomes per root section was counted in control conditions or after DTT and TM treatment as above and the average number determined for 20 seedlings per treatment. Error bars represent se. Insets show enlargement of indicated boxes. Previously, ATG18a was identified as an essential gene for autophagy induction under several abiotic stresses. Thus, RNA interference RNAi - ATG18a transgenic plants showed an autophagy-defective phenotype Xiong et al. To investigate whether ATG18a is required for ER stress—induced autophagy, 7-d-old RNAi- ATG18a seedlings were subjected to ER stress as described above Figure 1B. No autophagosomes were detected under control conditions or upon TM or DTT treatment; this indicates that ER stress—induced autophagy is dependent on the function of ATG18a and, therefore, most likely occurs via the classical autophagy pathway. There are two possibilities to explain the increased number of autophagosomes seen under ER stress; either the ER stress leads to an increased rate of formation of autophagosomes, or the ER stress inhibits the delivery to or degradation of autophagosomes in the vacuole. ConcA raises the vacuolar pH and inhibits trafficking of vacuolar proteins, preventing vacuolar protein degradation Matsuoka et al. Autophagic bodies therefore accumulate in vacuoles after treatment with concA instead of being degraded Yoshimoto et al. In control conditions, the GFP signal was diffuse and the vacuole rarely contained any spherical structures in the DIC images. After adding TM , several GFP-ATG8e—labeled puncta and spherical structures were observed, indicating the formation of autophagosomes. This indicates that after adding TM and concA , autophagic bodies accumulate inside the vacuole instead of being degraded. Identical results were obtained using DTT instead of TM see Supplemental Figure 1 online. Together, these results demonstrate that the increased number of autophagosomes present upon TM treatment is due to the increased formation of autophagosomes, rather than decreased degradation because of a deficiency in delivery to or fusion between autophagosomes and vacuoles. Autophagy is regulated by either NADPH oxidase—dependent or —independent pathways, depending on the stress conditions Liu et al. To determine whether ER stress—induced autophagy is dependent on NADPH oxidase, the NADPH oxidase inhibitors diphenyleneiodonium DPI and imidazole were used. Seven-day-old wild-type plants grown on MS plates were transferred to liquid MS medium plus DTT or TM , plus or minus imidazole or DPI , for 8 h Figure 1E. Compared with control conditions, wild-type plants had substantially more autophagosomes when treated with DTT or TM as shown above. This result implies that ER stress—induced autophagy is regulated by an NADPH oxidase—independent pathway. One role of autophagy during ER stress might be to degrade regions of the ER , thereby turning over some of the ER membrane and contents. To investigate the physical relationship between the autophagy pathway and the ER , the subcellular localization of both ER and autophagosome markers was analyzed in response to ER stress. GFP fused with the ER retention signal HDEL was used to label the ER Batoko et al. To prevent vacuolar degradation, 1 µM concA or DMSO as carrier control was added to the corresponding stress-inducing liquid MS medium, followed by incubation for 12 h in the dark. Seedlings were imaged by confocal microscopy, with two focal planes shown for each sample: one through the cortical cytoplasm and one through the vacuole. In the control, confocal microscopy showed typical ER networks in the cytoplasm, and the vacuole was relatively devoid of any GFP signal Figure 2 , left panels. Likewise, few spherical structures were observed in DIC images of the vacuole in the controls Figure 2 , right panels. The concA -treated sample also showed typical ER patterns in the cytoplasm and no GFP signal in the vacuole, although in some cells small numbers of spherical structures were observed in the DIC images. The Localization of GFP-HDEL Changes during ER Stress in Arabidopsis Roots. Seven-day-old GFP-HDEL transgenic plants were transferred to liquid MS medium with or without 1 µM concA for 12 h control conditions. DMSO was used as a solvent control for all experiments. The plants were then observed with both confocal microscopy left panels and DIC microscopy right panels. Far-right panels show enlarged boxes. To induce ER stress, 7-d-old GFP-HDEL transgenic Arabidopsis seedlings were treated with TM or DTT for 8 h. With either treatment, the typical ER structure was still observed in the cytoplasm, although some punctate structures were occasionally seen. Vacuoles were still largely devoid of any GFP signal, and spherical structures were rarely seen in vacuoles by DIC imaging, similar to the control. In the —Suc and —N conditions, the GFP signal also appeared as an ER network in the cytoplasm and the spherical structures were rarely seen in the vacuole. These observations suggest that ER is transported to the vacuole via an autophagy-like pathway during ER stress but not largely transported to the vacuole during nutrient starvation conditions. To test whether the spherical structures that deliver ER to the vacuole are autophagosomes, leaf protoplasts obtained from 4-week-old GFP-HDEL plants were transformed with a cerulean-ATG8e fusion construct, then incubated in the dark for 12 h to allow gene expression. Confocal microscopy was performed to visualize the subcellular localization of both GFP-HDEL and cerulean-ATG8e Figure 3A. Note: The plant protoplasts expressing only GFP-HDEL or cerulean-ATG8e displayed little fluorescence contamination between these two individual signals. In the controls, GFP-HDEL showed a typical cytoplasmic ER pattern, and the cerulean-ATG8e gave a diffuse cytosolic signal. In the presence of TM , both the cerulean- and the GFP -labeled puncta appeared in most of the cells, and some of the cerulean puncta and GFP puncta colocalized with each other. After incubation with both TM and ConcA, in most of the cells, numerous GFP puncta appeared as shown previously in planta, and the cerulean-ATG8e was also found in multiple puncta. Many of the structures labeled with these two markers colocalized, and in the bright-field images, these puncta corresponded to small vesicles in the vacuole. Autophagy Delivers ER to the Vacuole during ER Stress in Arabidopsis Leaf Protoplasts. A Leaf protoplasts obtained from 4-week-old GFP-HDEL plants were transformed with a cerulean-ATG8e fusion construct. For control conditions, the protoplasts were incubated in W5 solution with or without 1 µM concA for 12 h. Confocal microscopy was used to visualize the GFP and cerulean fluorescence. GFP-HDEL protoplasts lacking the cerulean-ATG8e construct were used as a GFP fluorescence control; wild-type protoplasts transformed with the cerulean-ATG8e construct were used as a cerulean fluorescence control. Arrows indicate autophagic bodies labeled with both GFP-HDEL and cerulean-ATG8e. C Leaf protoplasts obtained from 4-week-old wild-type WT or RNAi- ATG18a plants were transformed with a CFP-HDEL fusion construct. For the control, the protoplasts were incubated in W5 solution with or without 1 µM concA for 12 h. Confocal microscopy was used to visualize CFP fluorescence. Arrows indicate CFP-HDEL—labeled structures inside the vacuole. D Pearson's colocalization coefficient for GFP-HDEL and cerulean-ATG8e autophagosomes , CFP-HDEL and ST-GFP Golgi , CFP-HDEL and GFP-VHA1 TGN , and CFP-HDEL and YFP-RHA1 PVC. Pearson's coefficient was derived from three independent experiments. To analyze further and quantify the colocalization between the cerulean-ATG8e and the GFP-HDEL signals, the fluorescence patterns of the two signals were analyzed using the ImageJ software Abramoff et al. Together, these data suggest that during ER stress, ER is delivered to the vacuole through an ATG8e-containing vesicle, presumably an autophagosome. To test further the role of the autophagy pathway in delivering ER to the vacuole, similar experiments as described above were performed comparing wild-type and RNAi- ATG18a leaf protoplasts transiently expressing a cyan fluorescent protein CFP -HDEL fusion construct as an ER marker Liu et al. Autophagosome formation is defective in RNAi- ATG18a plants, which thus can be used to test whether the loss of the autophagy pathway blocks ER transport to the vacuole during ER stress. In both wild-type and RNAi- ATG18a protoplasts, the CFP signal labeled an ER membrane network in control conditions. After addition of concA , most of the wild-type protoplasts showed an ER pattern and a few showed some CFP puncta, but almost all RNAi- ATG18a protoplasts observed displayed an ER pattern without CFP puncta. After adding TM , CFP puncta were observed in a majority of wild-type protoplasts, but not in RNAi- ATG18a protoplasts. After adding both TM and concA to the medium, most of the wild-type protoplasts observed accumulated numerous CFP puncta inside the vacuole; however, RNAi- ATG18a protoplasts still displayed the ER pattern, suggesting that delivery of ER to the vacuole is blocked when autophagy is defective. These results indicate that the delivery of ER to the vacuole is dependent on the autophagy-related gene ATG18a and this, when taken together with the colocalization between the ER marker and the autophagosome marker, suggests that the ER is delivered in autophagosomes. Two major vacuolar trafficking routes have been identified in plants: the autophagy pathway and the biosynthetic pathway, which involves the Golgi, the trans -Golgi network TGN , and the prevacuolar compartment PVC Sanderfoot et al. To investigate whether the ER structures identified in the vacuole can also be transported through the biosynthetic pathway, the subcellular colocalization between the ER marker and the Golgi marker ST-GFP Boevink et al. Both the confocal images and the Pearson coefficient Figure 3D suggested that the punctate ER fluorescence signal does not colocalize with the Golgi, the TGN , and the PVC structures. These results indicate that the ER structures identified in the vacuole during ER stress are not transported via the Golgi, TGN , and the PVC pathway. Although it can be interpreted that the colocalization of ER puncta with an autophagosome marker indicates that ER is transported to the vacuole by autophagosomes, previous studies in animals showed that the autophagosome membrane can be derived from ER membrane Hayashi-Nishino et al. To clarify whether ER is delivered by autophagosomes or is a component of the autophagosome membrane during ER stress, electron microscopy was performed to examine the detailed structures of ER stress—induced autophagosomes Figure 4. In the controls, a few small vesicles were observed in both the vacuoles and the cytoplasm Figure 4A. In response to TM treatment, numerous vesicles appeared in the cytoplasm, but the contents of the vesicles were difficult to identify Figure 4C. This might be due to the fusion of autophagosomes with a smaller lysosome-like or endosome-like organelle, leading to the degradation of the contents in the autophagosomes before fusion with the vacuole Rose et al. Since concA was used to inhibit vacuolar degradation, the contents inside the autophagic bodies could be identified. These small vesicles contained a variety of cargos, some with unidentified cytoplasmic contents Figure 4H , whereas many had membrane structures decorated with electron-dense ribosomes Figures 4E to 4G , typical of ER. Not all autophagosomes or autophagic bodies contained ER , consistent with the partial colocalization seen by confocal microscopy. From this we conclude that ER membranes had been engulfed by autophagosomes. Together, these results imply that ER is transported to the vacuole for degradation via the autophagy pathway during ER stress. ER Membranes Are Engulfed by Autophagosomes during ER Stress. AB, autophagic bodies. E and F Autophagic bodies with ribosome-decorated membranes inside. G Enlargement of a section indicted in F. In Arabidopsis , IRE1 has been identified as an ER stress sensor. IRE1 senses ER stress and splices the mRNA encoding bZIP60, which is a bZIP -containing transcription factor implicated in the UPR in plants Deng et al. There are two members of the IRE1 gene family in Arabidopsis , IRE1a and IRE1b Koizumi et al. To investigate whether ER stress—induced autophagy is activated via IRE1 genes, the induction of autophagy was examined in ire1a or ire1b null mutants Humbert et al. In control conditions, all three genotypes showed very few autophagosomes. ER Stress—Induced Autophagy Is Dependent on IRE1b Function in Arabidopsis Roots. B The number of MDC -stained autophagosomes per root section was counted after DTT and TM treatment as above and the average number determined for 20 seedlings per treatment. To quantify these observations, autophagosome numbers were analyzed per root section for both ire1a and ire1b mutants Figure 5B. The ire1a mutant responded to both ER stress DTT or TM treatment and the starvation stress, whereas the ire1b mutant responded to the starvation stress but showed significantly lower autophagosome numbers during ER stress treatment. To confirm further the MDC staining result, concA was used to prevent vacuolar degradation of autophagic bodies, which were visualized by DIC microscopy. In the absence of concA , ire1a , ire1b , and wild-type plants all displayed few spherical structures in the vacuole in almost all conditions tested. The DIC image analysis together with the MDC staining results indicate that IRE1b , and not IRE1a , is required for ER stress—induced autophagy but not for starvation-induced autophagy in roots. To confirm further the MDC and DIC results obtained in seedling roots, 4-week-old leaf protoplasts from wild-type, ire1a , and ire1b plants were transformed with a GFP-ATG8e fusion construct to visualize the induction of autophagy under ER stress by confocal microscopy Figure 6A. In the control, the fluorescence from GFP-ATG8e was diffuse in all three types of plant protoplasts. In response to TM treatment, most of the wild-type and ire1a plants contained GFP-ATG8e labeled puncta, indicating the induction of autophagy, whereas the GFP signal was diffuse in ire1b , indicating no autophagosome formation. These results again suggest that the ER stress—induced autophagy is dependent on IRE1b. ER Stress—Induced Autophagy Is Dependent on IRE1b Function in Arabidopsis Leaf Protoplasts. Leaf protoplasts obtained from 4-week-old wild-type WT , ire1a , and ire1b plants were transformed with GFP-ATG8e A or CFP-HDEL B fusion constructs. As a control, protoplasts were incubated in W5 solution with or without 1 µM concA for 12 h. DMSO was used as a solvent control. Confocal microscopy was used to visualize the GFP and CFP fluorescence. Arrows indicate autophagic bodies containing GFP-ATG8e or CFP-HDEL. Next, the role of IRE1b in delivery of ER to the vacuole upon ER stress was analyzed using wild-type, ire1a , and ire1b leaf protoplasts transiently expressing a CFP-HDEL fusion construct Figure 6B. In control conditions, the ER pattern was typical in all three types of plant protoplasts. In the presence of TM , most of the wild-type and ire1a plants contained CFP -labeled puncta, whereas ire1b plants mainly showed a typical ER labeling pattern. These results imply that the delivery of ER to the vacuole is dependent on IRE1b but not on IRE1a. Together, the findings both in planta and in protoplasts suggest that IRE1b is required for ER stress—induced autophagy. To confirm that the loss of autophagy induction during ER stress in the ire1b mutant was actually due to the lack of IRE1b gene function, autophagy induction was tested in both ire1b leaf protoplasts transiently expressing a FLAG-tagged IRE1b construct Figure 7A and transgenic lines expressing the IRE1b cDNA IRE1b-FLAG in the ire1b mutant background Figure 7B ; see Supplemental Figure 6 online. Wild-type leaf protoplasts transiently expressing GFP-ATG8e displayed a diffuse GFP signal in control conditions, and GFP puncta were observed in the presence of TM as expected Figure 7A. ire1b leaf protoplasts transiently expressing GFP-ATG8e showed a diffuse GFP signal in both the control and after TM treatment. However, upon transformation of ire1b leaf protoplasts with both IRE1b-FLAG and GFP-ATG8e constructs, the GFP signal was diffuse in the control conditions, but GFP puncta were seen in the presence of TM , similar to wild-type protoplasts. Together, these results indicate that the defect for autophagy induction in ire1b in response to ER stress can be attributed to the loss of IRE1b gene function, rather than other defects in the autophagy pathway. This again suggests that ER stress—induced autophagy is dependent on the IRE1b gene. Defects in Autophagy Induction in ire1b during ER Stress Can Be Attributed to the Loss of IRE1b Gene Function. A Leaf protoplasts obtained from 4-week-old wild-type WT or ire1b plants were transformed with the GFP-ATG8e fusion construct, or ire1b leaf protoplasts were transformed with both GFP-ATG8e and IRE1b-FLAG fusion constructs. For controls, the protoplasts were incubated in W5 solution. Arrows indicate GFP-ATG8e—labeled autophagic bodies. Arrows indicate MDC -stained autophagosomes and autophagic bodies. As discussed above, IRE1b splices bZIP60 mRNA to produce an active transcription factor, thus upregulating the UPR genes in plants Deng et al. To test whether regulation of ER stress—induced autophagy by IRE1b occurs via IRE1b splicing of bZIP60 , the induction of autophagy was examined in a bzip60 T-DNA insertion mutant Deng et al. Seven-day-old wild-type and bzip60 plants grown on MS plates were transferred to MS liquid medium supplemented with TM or DSMO as a solvent control, followed by MDC staining Figure 8. Unexpectedly, the bzip60 mutant showed constitutive autophagy even under control conditions. One explanation for the constitutive autophagy in the bzip60 mutant is that the loss of bZIP60 function causes constitutive ER stress, thus inducing autophagy. Alternatively, the loss in bZIP60 function may lead to general cellular stress, causing an increased level of basal autophagy. This complicated the testing of whether ER stress induces autophagy in bzip60 , as autophagy seen upon TM treatment in bzip60 could either be increased basal autophagy or a mixture of the basal autophagy and TM induced autophagy. bZIP60 and bZIP28 Are Not Involved in Regulating ER Stress—Induced Autophagy. MDC staining of roots was performed to visualize autophagosomes. To distinguish between these two possibilities, the NADPH oxidase inhibitor DPI was used to inhibit the general starvation and salt stress—induced autophagy pathway Liu et al. As shown in Figure 1 , the addition of an NADPH oxidase inhibitor does not block ER stress—induced autophagy. In the presence of DPI , no autophagy was seen in bzip60 Figure 8 , indicating that the constitutive autophagy observed in bzip60 is inhibited by DPI and is therefore most likely a general stress response and unrelated to ER stress. After adding both DPI and TM to the medium, wild-type plants still showed autophagy induction. Autophagosomes were also present in the bzip60 mutant in the presence of DPI and TM , which suggests that after DPI inhibition of the enhanced basal autophagy, an alternative pathway for activation of ER stress—induced autophagy was still active. These data indicate that autophagy can still be induced by ER stress in the bzip60 mutant and, therefore, that bZIP60 is not required for ER stress—induced autophagy. Thus, ER stress—induced autophagy is regulated by IRE1b but is not dependent on the downstream factor bZIP Animal cells contain another two ER stress sensors, ATF6 and PERK , in addition to IRE1. Cells lacking ATF6 or PERK are capable of autophagy induction in response to ER stress Ogata et al. In plants, bZIP28 may be functionally equivalent to ATF6, whereas PERK signaling has not been demonstrated in plants Liu et al. To investigate whether bZIP28 is involved in ER stress—induced autophagy, a bzip knockout mutant Liu et al. Similar experiments as described above for bzip60 were performed with 7-d-old bzip plants Figure 8. The bzip plants displayed constitutive autophagy even in control conditions. This constitutive autophagy was inhibited by the addition of the NADPH oxidase inhibitor DPI , indicating that the constitutive autophagy seen in the bzip mutant was most likely a general stress response and unrelated to ER stress. After adding both DPI and TM to the medium, autophagosomes were present in bzip , indicating that after the inhibition of general stress-induced autophagy, autophagy can still be induced by ER stress in the bzip mutant. These data imply that, like bZIP60 , bZIP28 is not required for ER stress—induced autophagy. Although a number of studies have focused on UPR signaling pathways in plants, little is understood about ER morphology changes in response to ER stress or as mediated by the UPS -independent ERAD pathway Urade, , ; Moreno and Orellana, Previously, plant autophagy had been shown to be involved in senescence, nutrient deprivation, oxidative stress, salt and drought stresses, and pathogen infection Doelling et al. In this article, we demonstrate that autophagy is activated in the response to ER stress in plants. MDC staining and GFP-ATG8e transgenic plants showed autophagy induction after TM or DTT treatment. In addition, portions of the ER are engulfed by autophagosomes and delivered to the vacuole for degradation. Together, this evidence implicates autophagy in ER turnover in response to ER stress. To investigate the upstream signaling pathway that activates ER stress—induced autophagy, a mutant lacking one of the ER stress sensors, IRE1b , was tested for autophagy induction upon ER stress. Leaf protoplasts transiently expressing CFP-HDEL or GFP-ATG8e indicated that IRE1b is required for ER stress—induced autophagy. To characterize further the IRE1b-dependent autophagy pathway, a mutant lacking the splicing target of IRE1b, bZIP60, was also analyzed. The bzip60 mutant was capable of inducing autophagy in response to ER stress, suggesting that ER stress—induced autophagy does not rely on the splicing activity of IRE1b. Our data identified IRE1b as an upstream component of ER stress—induced autophagy in Arabidopsis seedlings. However, we cannot exclude the possibility that IRE1a could also be involved in autophagy. Thus, by analyzing root tissues and protoplasts, we may not have been able to assess the contribution by IRE1a simply because it is not highly expressed in roots. IRE1a plays newly recognized roles in plant defense responses, so it will be interesting to determine whether those responses also involve autophagy Moreno et al. There is some discrepancy in the literature about the extent to which the roles of IRE1a and IRE1b overlap, which may be due to allelic differences in the mutants used in the different studies Deng et al. The detailed molecular mechanism of regulation of ER stress—induced autophagy is yet to be determined. In yeast, ER stress—induced autophagy is regulated through the IRE1 endoribonuclease activity toward HAC1 mRNA Yorimitsu et al. In animals, IRE1 is also required for autophagy induction; however, the IRE1 kinase activity-mediated c-Jun N-terminal kinase pathway, which is absent in plants, rather than the splicing activity toward XBP1 seems to control autophagy induction Urano et al. Our results showed that in plants, ER stress—induced autophagy is dependent on IRE1b , suggesting a conserved role for the IRE1 gene during autophagy induction from yeast to animals and plants. Similar to animals, autophagy does not depend on the IRE1b downstream splicing target, which in the case of Arabidopsis is bZIP60 Deng et al. As bZIP60 is the only known target of IRE1b ribonuclease activity, this suggests that either 1 IRE1b has additional splicing targets that have yet to be discovered that regulate autophagy activation or 2 other activities associated with IRE1b may be responsible for the autophagy induction in response to ER stress, rather than its splicing activity. However, other functions of IRE1b in addition to the splicing of bZIP60 mRNA have not been discovered to date Deng et al. This suggests that a distinct, previously undiscovered, signaling pathway functions in activation of autophagy during ER stress in Arabidopsis. To elucidate further the role of IRE1 in autophagy induction, more experiments are needed to identify its substrates and downstream signals. Intriguingly, the loss of XBP1 in Drosophila melanogaster causes constitutive autophagy Arsham and Neufeld, , similar to that of the bzip60 and bzip28 mutants observed here. The authors suggest that the absence of XBP1 activity may lead to accumulation of unfolded proteins, triggering XBP1-independent UPR signaling. Whether this happens in plant cells still needs to be determined. Target of rapamycin TOR has been shown to be a negative regulator of autophagy from yeast to animals and plants Díaz-Troya et al. TOR regulates the downstream ATG1 kinase complex, recently characterized in Arabidopsis Suttangkakul et al. Several studies have shown an interplay between ER stress and mTOR signaling in animals; for example, constitutive activation of mTOR leads to ER stress Ozcan et al. It was also suggested that ER stress induces autophagy through the inactivation of mTOR Qin et al. However, whether TOR is associated with the control of autophagy during ER stress in plants is still unknown. A recent study in Chlamydomonas reinhardtii reported that the phosphorylation state of the BiP chaperone is regulated by TOR Díaz-Troya et al. The authors showed that under ER stress when increased chaperone levels are needed, BiP protein is dephosphorylated, resulting in its activation. When protein synthesis was inhibited by downregulating TOR activity, BiP was phosphorylated to its inactive form Díaz-Troya et al. These results indicate a potential TOR function in its interaction with the ER stress signal, thereby regulating both protein synthesis and the autophagy degradation pathway. However, how exactly TOR senses ER stress, and whether IRE1 fits into this pathway, still needs to be determined. Generally, autophagy is a nonselective process; however, organelle-specific autophagy has been identified in both yeast and animals Reumann et al. For example, the selective degradation of peroxisomes pexophagy Hutchins et al. In plants, organelle-specific autophagy has not been studied extensively. Nevertheless, increasing evidence is emerging for organelle-specific autophagy in plants, such as the degradation of ribosomes Hillwig et al. However, whether the engulfment of ER by autophagosomes is a selective process is unknown. One possibility is that during ER stress, the ER begins to fragment, allowing it to be incorporated into autophagosomes nonselectively. Another possibility is that the autophagosome can recognize ER fragments containing misfolded proteins Yorimitsu and Klionsky, , therefore sequestering both the misfolded proteins and the ER membranes. Similar mechanisms have been reported in plants. NBR1 homologs in both Arabidopsis and tobacco Nicotiana tabacum have been identified, and they both interact with ATG8 Svenning et al. Therefore, it is possible that the autophagosome sequesters ER through identifying ER fragments containing protein aggregates or containing surface proteins tagged by ubiquitylation, as has been seen in the case of mitophagy Ashrafi and Schwarz, ; however, more evidence is required before this conclusion can be drawn. In this study, we provide another link between organelle degradation and autophagy by showing that the ER is a target of autophagy during ER stress in plants. Activities other than the splicing of bZIP60 by IRE1b may function as upstream events to regulate ER stress—induced autophagy. However, future experiments are needed to determine the downstream targets of IRE1b and the detailed regulation mechanisms in the ER stress—induced autophagy pathway. Arabidopsis thaliana Columbia ecotype seeds were surface sterilized with 0. Transgenic plants used in this study have been described previously as follows: RNAi- ATG18a Xiong et al. For starvation treatment, 7-d-old seedlings grown on solid MS plates were transferred to MS plates lacking Suc or nitrogen for an additional 4 d. Plants grown on Suc starvation plates were incubated in the dark. If concA treatment see below was also required, the seedlings were then transferred to liquid MS medium lacking Suc or nitrogen plus concA for 12 h in the dark. For imidazole and DPI treatment, seedlings grown on solid MS plates were transferred to MS liquid medium plus or minus 20 mM imidazole or 20 µM DPI for the indicated times. The solvent for DPI was DMSO; an equivalent volume of DMSO was added to controls. For concA treatment, seedlings grown on MS plates were transferred to MS liquid medium containing 1 µM concA or DMSO as a solvent control for 12 h in the dark. The roots were mounted in water and then observed by confocal, fluorescence, and DIC microscopy. Arabidopsis seedlings were stained with MDC as previously described Contento et al. Seedlings were incubated with 0. Confocal microscopy was performed with a Leica confocal microscope using a ×63 Leica oil immersion objective. The ATG8e cDNA was synthesized by RT-PCR from total RNA from 7-d-old seedlings grown on MS plates, using gene-specific primers see Supplemental Table 1 online. The cDNA was sequenced for verification and ligated into the pAN vector using Bgl II and Not I restriction sites Rizzo et al. Protoplasts transformed with the Cerulean-ATG8e construct were observed with a CFP -optimized filter. Arabidopsis leaf protoplasts were prepared and transformed according to Sheen Twenty micrograms of plasmid DNA was used for each transformation. Protoplasts were incubated at room temperature in darkness for 12 h, with 40 rpm orbital shaking. Pearson's colocalization coefficients were derived using ImageJ software Abramoff et al. All Pearson's coefficients were derived from three completely independent experiments. IRE1b coding sequence was amplified from Columbia-0 cDNA using gene-specific primers. A 3XFLAG tag was added after the transmembrane domain of IRE1b by overlapping PCR. Primers used are listed in Supplemental Table 1 online. IRE1b-N primers were used to amplify the first half of the IRE1b gene up to and including the transmembrane domain, IRE1b-C primers were used to amplify the second half of the IRE1b gene after the transmembrane domain, and FLAG primers were used for the 3XFLAG tag. The IRE1b-3XFLAG DNA fragments were then ligated into the pSKM36 vector using Asc I- Spe I restriction sites Ikeda et al. This construct was introduced into Agrobacterium tumefaciens by electroporation Mersereau et al. Transgenic plants were identified by kanamycin resistance. Individuals from the T2 generation were used for further studies. Electron microscopy was performed at the Iowa State University Microscopy and NanoImaging Facility. Samples were rinsed three times in 0. Resin blocks were polymerized for 48 h at 65°C. Images were captured using a JEOL scanning and transmission electron microscope Japan Electron Optic Laboratories. Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At2g IRE1a , At5g IRE1b , At3g bZIP28 , At1g bZIP60 , and At3g ATG18a. The following materials are available in the online version of this article. Supplemental Figure 1. Autophagy Is Activated in the Presence of DTT. Supplemental Figure 2. CFP-HDEL Does Not Colocalize with Golgi Structures during ER Stress. Supplemental Figure 3. CFP-HDEL Does Not Colocalize with a TGN Marker during ER Stress. Supplemental Figure 4. CFP-HDEL Does Not Colocalize with a PVC Marker during ER Stress. Supplemental Figure 5. Autophagosomes Do Not Accumulate in ire1b Roots in Response to ER Stress. Supplemental Figure 6. Supplemental Table 1. Primers Used for Generation of the Cerulean-ATG8e and IRE1b-FLAG Constructs and for PCR. We thank Harry Jack T. Horner, Randall Den Adel, and Tracey M. Pepper for assistance with the fluorescence, DIC , and the electron microscopy and Margaret Carter for assistance with confocal microscopy. We also thank Ian Moore for the GFP-HDEL transgenic plant, Karin Schumacher, Chris Hawes, and Erik Nielsen for constructs, and Anthony L. Contento for generating the cerulean-ATG8e construct. This research was supported by Grants IOB and MCB from the National Science Foundation to D. designed research. performed research. analyzed data. wrote the article. Abramoff M. Magalhaes P. Ram S. Image Processing with ImageJ. Biophotonics International 11 : 36 — Google Scholar. Arsham A. Neufeld T. A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway. PLoS ONE 4 : e Ashrafi G. Schwarz T. June 29, The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. Bassham D. Plant autophagy—More than a starvation response. Plant Biol. Batoko H. Zheng H. Hawes C. Moore I. A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12 : — Bernales S. McDonald K. Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. Boevink P. Oparka K. Santa Cruz S. Martin B. Betteridge A. Plant J. Calfon M. Zeng H. Urano F. Till J. Hubbard S. Harding H. Clark S. Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature : 92 — Che P. Bussell J. Zhou W. Estavillo G. Pogson B. Smith S. Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Chen Y. Brandizzi F. Clough S. Bent A. Floral dip: A simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Contento A. Xiong Y. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Cox J. Shamu C. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73 : — A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87 : — Deng Y. Humbert S. Liu J. Srivastava R. Rothstein S. Howell S. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. USA : — Dettmer J. Hong-Hermesdorf A. Stierhof Y. Schumacher K. Plant Cell 18 : — Díaz-Troya S. Pérez-Pérez M. Florencio F. Crespo J. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4 : — Pérez-Martín M. Moes S. Jeno P. Inhibition of protein synthesis by TOR inactivation revealed a conserved regulatory mechanism of the BiP chaperone in Chlamydomonas. Plant Physiol. Doelling J. Walker J. Friedman E. Thompson A. Vierstra R. Dröse S. Bindseil K. Bowman E. Siebers A. Zeeck A. Altendorf K. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32 : — Gardner B. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science : — Hanaoka H. Noda T. Shirano Y. Kato T. Hayashi H. Shibata D. Tabata S. Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Zhang Y. Bertolotti A. Perk is essential for translational regulation and cell survival during the unfolded protein response. Cell 5 : — Hayashi-Nishino M. Fujita N. Yamaguchi A. Yoshimori T. Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Cell Biol. Haze K. Yoshida H. |
| Endoplasmic Reticulum Stress and Dysregulated Autophagy in Human Pancreatic Beta Cells | To test Skinfold measurement in bodybuilding the spherical structures that Auhophagy ER strress the vacuole are autophagosomes, leaf Autophagy and ER stress atress from 4-week-old GFP-HDEL plants were transformed with a strews fusion construct, then ajd Autophagy and ER stress the dark for 12 h to allow gene expression. Simultaneously, the ATF6α-mediated upregulation of CHOP, XBP1, and GRP78 expression is also initiated, resulting in the induction of autophagy [ ]. Activation of the PERK pathway contributes to expression of multiple ATG genes Cai et al. Confocal and electron microscopy showed that portions of the ER are engulfed by autophagosomes and delivered to the vacuole, most likely for degradation. Kraft C. |
| The Role of Autophagy and ER Stress in Viral Infection | Yamane T. Kirkin V. Life Sci. Strexs, T. Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M et al. Zientara-Rytter K. |
| REVIEW article | PERP is therefore likely to be Autophgay Autophagy and ER stress molecule that drives damaged Autolhagy towards apoptosis 7. However, Muscle recovery foods the engulfment of ER by autophagosomes is a selective process is unknown. DOWNLOAD Autophagy and ER stress FREE Strwss Cite Cite this chapter There are two ways to cite this chapter:. Linking ER stress to autophagy: potential implications for cancer therapy. Figure 2. The dissociation of GRP78 drives ATF6 to translocate to the Golgi, where it is cleaved by site-1 protease S1P and S2P to generate ATF6f, which transcriptionally activates the expression of a variety of genes involved in ERAD and ER chaperones including GRP78 and XBP1 Haze et al. Kraft C. |

Autophagy and ER stress -
In addition, during ER stress, formed ATF6f induces the expression of DAPK1 Kalvakolanu and Gade, ; Gade et al. Dopaminergic neurons are particularly sensitive to unfolded, misfolded, and excessively aggregated proteins. ER stress and autophagy impairment are two essential events that lead to the imbalance of proteostasis, which contributes to DA neurodegeneration.
In recent years, a large number of studies have focused on the relationship between ER stress and PD pathogenesis.
For example, injection of an ER stress inducer, tunicamycin, into mouse brains causes high levels of oligomeric α-SYN, DA neuron death, locomotor deficiency, and glial activation Coppola-Segovia et al. However, impaired autophagy, CMA, and mitophagy are frequently observed rather than autophagy activation in PD.
Interestingly, conditional deletion of the Atg7 gene in mice recapitulates many of the pathologic features of PD, including age-related loss of DA neurons, loss of striatal DA, accumulation of α-SYN, and ubiquitinated protein aggregates Ahmed et al.
Thus far, it is generally recognized that in PD patients as well as various PD cellular and animal models, ER stress activation, UPR markers, and autophagy dysfunction undoubtedly exist in the lesion regions, and are closely related to both genetic and neurotoxic factors that induce DA neurodegeneration.
The UPR activation markers, including phosphorylation of PERK, eIF2α, and IRE1α, are observed in neuromelanin-containing DA neurons in the postmortem SNpc of PD patients rather than age-matched controls Hoozemans et al.
In addition, the immunoreactivity of phosphorylated PERK is co-localized with increased α-SYN immunoreactivity in DA neurons Hoozemans et al.
Importantly, UPR activation is an early event in neurodegeneration and is closely associated with the accumulation and aggregation of α-SYN Hoozemans et al. GRP78 and CHOP, which are ER stress markers, are increased in the SNpc in PD patients Selvaraj et al.
Moreover, GRP78 is increased to a greater extent in dementia with LB DLB and PD with dementia PDD patients in the cingulate gyrus and parietal cortex Baek et al.
GRP78 is also dramatically upregulated in the brain tissues of autosomal recessive juvenile PD AR-JP patients caused by a loss of functional Parkin, a familial PD genetic factor Imai et al.
The protein disulfide isomerase PDI family participates in disulfide bond formation, reduction, isomerization, and accurate folding of nascent proteins, and they also are increased to constitute an adaptive response to ER stress Turano et al.
It was found that a PDI member, pancreatic PDI PDIp , accumulates in PD patient tissues Conn et al. Homocysteine-induced ER protein HERP is a stress response protein that functions in ER folding and ERAD-mediated degradation, and ER load reduction is also increased in the SN of PD patients Slodzinski et al.
However, all PD patients do not experience an increase in the UPR in tissues. Baek et al. Recently, similar findings suggested that both GRP78 and ATF4 protein levels are decreased in the SNpc in PD patients Esteves and Cardoso, These inconsistent results may be related to the different pathological degrees of PD patients.
In the early stage of PD, UPR activation is an adaptive response to protect DA neurons from damage. However, in the late stage, excessive stress-induced neuron damage or severe DA neuron loss leads to inhibition of expression of these ER stress markers.
Finally, ER stress and UPR activation are undoubted pathological processes in PD. It was first discovered that autophagic degeneration of DA neurons occurred in the SN regions of PD patients in early studies Anglade et al.
Later, immunopositivity for LC3-II indicated that autophagosome formation occurred in the majority of LBs and LNs, and LC3-II colocalized with α-SYN in PD patients Alvarez-Erviti et al. Moreover, enlarged mitochondria have been observed within autophagosomes using confocal laser scanning microscopy in PD brains, suggesting PD-associated abnormal mitophagy Zhu et al.
Fiesel and Springer ; Fiesel et al. Levels of HSC70 and LAMP2A were also dramatically decreased, which indicated that the CMA activity is significantly reduced in the SN tissues of PD patients Alvarez-Erviti et al.
The selective loss of LAMP2A protein and decreased levels of HSC70 were directly correlated with the increase in α-SYN levels and the accumulation of cytosolic CMA substrate proteins in PD samples Murphy et al.
Interestingly, crowded organelles and lipid membranes, including dystrophic lysosomes, mitochondria, and autophagosome-like structures, were observed in a recent PD postmortem study Shahmoradian et al.
Together, impairments in autophagy and CMA have been found in lesion regions in PD patients compared with matched controls using postmortem tissues. The increased autophagosomes and failed lysosomal clearance are common hallmarks in SN tissues of PD patients. The activated ER stress and UPR pathway may affect increases in autophagosome formation through the mechanisms described above.
α-SYN is encoded by the SNCA gene, the mutations of which such as A53T, or duplication or triplication, have been linked to autosomal-dominant forms of PD. Aggregated α-SYN, especially the accumulation in the brain of its soluble oligomers, is one of most important causative factors for both hereditary and sporadic PD.
α-SYN is a major component deposited in LBs and LNs, in which it harbors extensive phosphorylation at Ser, which mediates its aggregation and toxicity Wang et al.
α-SYN can be degraded via multiple clearance machineries, including autophagy, CMA, and the ubiquitin proteasome system Hou et al.
Although wild-type, mutant, phosphorylated, and oligomeric α-SYN activate ER stress and promote autophagy induction, they block autophagic flux by impairing autophagosome maturation, fusion with lysosomes, and lysosomal biogenesis or functions Figure 4.
Figure 4. Cross-links between ER stress and autophagy in α-SYN-mediated pathology. Accumulated α-SYN binds to GRP78 and activates ER stress. α-SYN also induces ER stress by binding to ATF6 and inhibiting its translocation to Golgi bodies. Additionally, wild-type, mutant, or phosphorylated α-SYN activates ER stress by inhibiting ER-Golgi trafficking, which leads to the accumulation of aggregated proteins.
The expression of BECLIN1 and LC3 triggers autophagy induction. However, wild-type and mutant α-SYN inhibit autophagosome maturation by repressing RAB1A function. They also activate mTORC1 and sequester TFEB in the cytoplasm to block autophagic flux by impairing lysosomal biogenesis and function.
α-SYN overexpression and its aggregated neurotoxic forms activate all three UPR branches and trigger chronic ER stress-induced apoptosis. It was discovered that overexpression of both wild-type and A53T mutant α-SYN affects RAB1, which is involved in trafficking substrates from the ER to the Golgi bodies, thus inducing UPR activation by blocking ER-Golgi trafficking.
RAB1 overexpression reduces stress and protects against DA neurodegeneration in PD animal models Cooper et al. A30P α-SYN disrupts the Golgi morphology and facilitates the susceptibility to ER stress.
A53T α-SYN upregulates GRP78 levels and eIF2α phosphorylation, and results in mitochondrial cell death in neurons, as well as in astrocytes Smith et al.
Accumulation of α-SYN within the ER activates the PERK pathway by directly interacting with GRP78 in vitro and in vivo Bellucci et al.
α-SYN oligomers rather than monomers also activate the IRE1α-XBP1 pathway Castillo-Carranza et al. In addition, α-SYN reduces ATF6 processing and leads to ERAD impairment by directly binding to ATF6 or indirectly restricting its incorporation into coat protein complex II COPII vesicles Credle et al.
ER stress also leads to the accumulation of α-SYN oligomers Jiang et al. It is interesting to hypothesize whether α-SYN toxicity is a cause or a consequence of ER stress and UPR dysfunction.
Additional evidence indicates that α-SYN accumulation within the ER is required for UPR activation, and toxic α-SYN oligomer formation precedes ER stress and UPR activation Colla et al. Nevertheless, collaboration of α-SYN accumulation-induced ER stress and ER stress-enhanced α-SYN neurotoxicity is vital for PD pathogenesis.
Both wild-type and A53T mutant α-SYN can promote autophagy induction by upregulating BECLIN1 and LC3 expression Yu et al. However, wild-type α-SYN overexpression impairs autophagic flux via RAB1A inhibition, leads to ATG9 mislocalization, and inhibits the formation of autophagosomes Winslow et al.
RAB1 overexpression protects DA neurodegeneration in various PD animal models Cooper et al. Additionally, α-SYN binds to high mobility group box 1 HMGB1 and strengthens BECLIN-BCL2 binding by blocking HMGB1-BECLIN1 interaction Song et al.
Both wild-type and A53T α-SYN can induce mTOR activity and impair autophagy Jiang et al. In an AAV-mediated α-SYN overexpression mouse model, α-SYN impairs autophagic efflux by sequestering TFEB in the cytoplasm, accompanied by p62 and LC3-II accumulation Decressac et al.
Similarly, PC12 cells harboring A53T α-SYN display accumulated autophagic-vesicular structures and impaired lysosomal hydrolysis Stefanis et al.
Interestingly, wild-type α-SYN contains a KFERQ sequence and is greatly degraded through the CMA pathway, while pathogenic α-SYN mutants act as CMA uptake inhibitors through interacting with LAMP2A on the lysosomal membrane Cuervo et al.
AAV-mediated overexpression of A53T α-SYN in neurons or A53T α-SYN in transgenic mice results in a reduction of CMA-mediated proteolysis of other substrates Xilouri et al. Interestingly, modification of wild-type α-SYN by DA also impairs CMA proteolysis, similar to mutant α-SYN Martinez-Vicente et al.
Whether wild-type and pathogenic α-SYN have an effect on phosphorylation and the activity of LAMP2A is not clear. Mutations in Parkin Kitada et al. Parkin is an E3 ligase and functions in the ERAD of misfolded ER proteins Shimura et al.
Parkin is upregulated by ATF4 under either ER stress or mitochondrial stress and inhibits stress-induced mitochondrial dysfunction and cell death via its E3 ligase activity Imai et al.
Conversely, an accumulation of PAEL receptor, a substrate of Parkin, induces ER stress and cell death Imai et al.
Furthermore, the ER stress inhibitor salubrinal prevents rotenone-induced ER stress and cell death through the ATF4-Parkin pathway Wu et al. Parkin also regulates ER stress and UPR via transcription factor pdependent XBP1 transcription regulation Duplan et al.
It has been reported that Parkin participates in ER stress regulation in DA neurons and also in astrocytes Ledesma et al. Moreover, an activation of the PERK branch of the UPR was observed that was induced by defective mitochondria, and PERK inhibition is neuroprotective in parkin mutant flies Celardo et al.
It has been shown that PINK1 inhibits ER stress-induced damage to mouse primary cortical neurons Li and Hu, , and downregulation of ER stress response genes has been detected in aged Pink1 knockout mice Torres-Odio et al.
Parkin and PINK1 are two important regulators that control mitophagy and mitochondrial homeostasis Swerdlow and Wilkins, Mitophagy, the process of removing damaged mitochondria, is compromised in PD pathogenesis, and its dysfunction is closely associated with DA neurodegeneration Liu et al.
It has been reported that the ER stress induced by tunicamycin TM and thapsigargin TG prevents Parkin loss and promotes its recruitment to the mitochondria, and also activates mitophagy during reperfusion after ischemia Zhang et al. However, the crosslink and mechanisms between ER stress and mitophagy in PD pathogenesis are mainly unknown.
LRRK2 possesses kinase function for catalyzing substrates and GTPase function for GTP-GDP hydrolysis. LRRK2 is co-localized with ER markers in DA neurons Vitte et al. LRRK2 phosphorylates leucyl-tRNA synthetase LRS , and this increases the number of misfolded proteins, causes ER stress, and induces autophagy initiation Ho et al.
LRRK2-GS mutation exacerbates these processes. It has recently been reported that LRRK2 regulates ER-mitochondria tethering through the PERK-mediated activation of E3 ligases, and LRKK2 mutation enhances the sensitivity to ER stress and decreases mitochondrial biogenesis Toyofuku et al. It has been reported that LRRK2 functions in endosomal- and vesicle-trafficking pathways, plays roles in cytoskeleton dynamics and neurite outgrowth, and regulates multiple steps of the autophagy-lysosome pathway Madureira et al.
Although wild-type and mutant LRRK2 promote autophagy by ER stress induced by the phosphorylation of LRS, they impair the autophagic degradation in an LRS-independent manner Ho et al.
LRKK2-GS fibroblasts exhibit higher autophagic activity levels involved in activating ERK activity rather than the mTOR pathway Bravo-San Pedro et al. LRRK2 and LRRK2-GS also regulate p62 phosphorylation, influence its affinity to ubiquitinated cargo Park et al.
Additionally, LRKK2 and LRKK2-GS inhibit autophagosome formation and autophagosome-lysosome fusion in various LRRK2-related PD models may through regulating phosphorylation of a number of RAB proteins Madureira et al.
LRKK2 pathogenic mutants also impair lysosomal function, which is detected by abnormal lysosomal morphology, abnormal cellular lysosomal localization, increased lysosomal pH, or inhibition of lysosomal enzymes Madureira et al. Like α-SYN, LRKK2 can also be degraded by CMA, and unlike α-SYN mutants, which increase the binding affinity of HSC70 and LAMP2A, LRRK2 mutants block the formation of the CMA translocation complex by inducing LAMP2A and HSC70 accumulation at the lysosomal membrane Orenstein et al.
Together, the promotion of autophagy initiation by LRRK2 and its pathogenic mutants is partly due to ER stress and UPR activation. However, they most likely inhibit autophagic flux, as well as CMA and lysosomal functions in an ER stress-independent manner.
DJ-1, a protein encoded by the PARK7 gene, assumes multiple functions including antioxidative stress and chaperone properties Mencke et al. Mutations or deletions of DJ-1 are associated with autosomal-recessive early-onset forms of PD Bonifati et al.
DJ-1 regulates ER stress and UPR by binding to and stabilizing ATF4 mRNA under both basal and stress conditions Yang et al. DJ-1 can also protect against ER stress-induced cell death in Neuro2a cells Yokota et al.
Moreover, it has been shown that oxidized DJ-1 can interact with N-terminal arginylated GPR78, and thus facilitate the self-polymerization of p62 and the targeting of pcargo complexes to phagophores under oxidative stress Lee et al. Interestingly, DJ-1 expression is regulated under ER stress such that XBP1 directly binds to its promoter and stimulates its expression Duplan et al.
Overexpression of DJ-1 in DA neurons and in the SN of rat brains promotes ERK-dependent autophagy. Although there are no obvious effects of DJ-1 deficiency on autophagy in SH-SY5Y cells Gonzalez-Polo et al.
In addition, DJ-1 deficiency in microglia impairs autophagy-mediated p62 degradation and reduces microglial-mediated α-SYN phagocytosis Nash et al. DJ-1 protects against DA neurodegeneration through enhancing CMA in PD animal models, SH-SY5Y cells, and astrocytes Xu et al.
Together, the upregulation of DJ-1 expression in response to ER stress may enhance the CMA or autophagic degradation of aggregated proteins, which bridges the close link between ER stress and autophagy in PD pathogenesis.
MPTP is the best-known chemical for inducing a PD model in vivo. Intraperitoneally injecting mice with MPTP reproduces PD pathology, including the selective loss of DA neurons in the SN and accumulation of protein aggregates, and eventually leads to the onset of PD-like clinical symptoms Bove et al.
MPTP also promotes the phosphorylation of p38 and enhances the interaction between phosphorylated p38 and ATF6, leading to an increase in ATF6 transcriptional activity Egawa et al. Increased levels of GRP78 and CHOP expression, as well as phosphorylated PERK and eIF2α, are detected in DA cellular models that are subjected to 6-OHDA treatment Ryu et al.
A recent study indicated that 6-OHDA treatment induces excessive autophagy with increased AMPK activity, decreased mTOR activity, reduced p62 levels, and also prevents alterations in lysosomal functions Chung et al.
In addition, 6-OHDA treatment stimulates CMA activity by increasing LAMP2A levels Wang et al. Together, 6-OHDA-induced excessive autophagy activation and autophagic flux contribute to PD pathogenesis.
Rotenone treatment produces most of the movement disorder symptoms and the histopathological features of PD, including LBs Betarbet et al. Rotenone also triggers ATF4 and CHOP expression involving activation of IRE1α and PERK in cellular models Ryu et al.
An induction of the IRE1α and PERK branch of the UPR has also been shown in rotenone rat or mouse models of PD Tong et al.
Treatment of N2a cells with rotenone triggers ER stress and the UPR involving all three branches of PERK, IRE1α, and ATF6 Gupta et al. Rotenone induces an increase in autophagy related proteins LC3-II and BECLIN1, as well as in autophagy substrates such as α-SYN and p62 in cultured PC12 cells Wu et al.
Rotenone increases oligomeric wild-type and A53T α-SYN in transfected cells through inhibiting their autophagic degradation Yu et al.
Therefore, rotenone-induced ER stress and the UPR initiate autophagy induction but block autophagic flux by impairing lysosomal functions, which aggravates the imbalance of cellular homeostasis and damage to DA neurons.
The accumulation of unfolded, misfolded, and aggregated proteins, and the accumulation induced by cellular stress are essential mechanisms underlying the causes of PD.
In this review, we systematically examined the intrinsic molecular links between ER stress, the UPR, and autophagy, as well as the roles of these cross-links in PD pathology. ER stress, UPR activation, and dysregulated autophagy commonly coexist in patients and various cellular and animal models of PD, and are closely related to DA neurodegeneration caused by PD genetic and neurotoxic factors Table 1.
This is why targeting one of these processes would create a beneficial PD treatment Moors et al. Table 1. The roles and mechanisms of PD-related factors in ER stress, autophagy and their cross-links. Figure 5. Proposed model of cross-links between ER stress, autophagy, and DA neurodegeneration.
PD-associated genetic and environmental factors trigger ER stress, and ER stress activates the UPR and induces autophagy to alleviate cellular stress. However, these PD-associated factors commonly block autophagic flux and impair lysosomal functions, and these changes synergistically cause severe damage and degeneration of DA neurons.
For example, administration of GSK, a PERK inhibitor, results in effective neuroprotection and prevents loss of SNpc DA neurons in mice that were treated with PD neurotoxin 6-OHDA Mercado et al. Gene therapy that restores the folding capacity by administration of viral-mediated overexpression of GRP78 Gorbatyuk et al.
Rapamycin, an inhibitor of mTOR, initiates autophagy induction, enhances autophagic flux Rubinsztein and Nixon, , and confers significant protective effects on DA neurons in various PD models Moors et al. Additionally, enhancing lysosomal biogenesis by TFEB overexpression or pharmacological stimulation of TFEB function by CCI was shown to eliminate α-SYN oligomers and rescue midbrain DA neurons from α-SYN toxicity in rats Decressac et al.
It is notable that PD-associated genetic or environmental factors lead to ER stress and UPR activation, which commonly initiate autophagy. However, these PD-associated factors also block autophagic flux and impair lysosomal functions. An intervention strategy for one of the two processes alone may not completely alleviate the imbalance in cellular homeostasis.
GW and XL designed the theme of the manuscript. HR and WZ wrote the manuscript. All authors approved the submitted version. This research was funded by the National Natural Science Foundation of China , , and , Suzhou Clinical Research Center of Neurological Disease Szzx , and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Abdullah, A. The unknown face of IRE1alpha - beyond ER stress. Cell Biol. doi: PubMed Abstract CrossRef Full Text Google Scholar.
Ahmed, I. Alvarez-Erviti, L. Chaperone-mediated autophagy markers in Parkinson disease brains. Anglade, P. Google Scholar. Arai, K. Involvement of ERK MAP kinase in endoplasmic reticulum stress in SH-SY5Y human neuroblastoma cells.
Arsikin, K. Autophagy-dependent and -independent involvement of AMP-activated protein kinase in 6-hydroxydopamine toxicity to SH-SY5Y neuroblastoma cells.
Acta , — Avivar-Valderas, A. Regulation of autophagy during ECM detachment is linked to a selective inhibition of mTORC1 by PERK.
Oncogene 32, — Baek, J. Unfolded protein response is activated in Lewy body dementias. Nucleic Acids Res. Bellucci, A. Bernales, S. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response.
PLoS Biol. Betarbet, R. Bioessays 24, — Bonifati, V. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science , — Bouman, L. Parkin is transcriptionally regulated by ATF4: evidence for an interconnection between mitochondrial stress and ER stress.
Cell Death Differ. Bove, J. NeuroRx 2, — Bozic, M. A conserved ATG2-GABARAP family interaction is critical for phagophore formation.
EMBO Rep. Bravo-San Pedro, J. Life Sci. Bruning, A. Nelfinavir and bortezomib inhibit mTOR activity via ATF4-mediated sestrin-2 regulation. Cai, Y. Interplay of endoplasmic reticulum stress and autophagy in neurodegenerative disorders.
Autophagy 12, — Castillo-Carranza, D. Differential activation of the ER stress factor XBP1 by oligomeric assemblies. Celardo, I.
Cell Death Dis. Chung, Y. Dysregulated autophagy contributes to caspase-dependent neuronal apoptosis. Colla, E. Endoplasmic reticulum stress is important for the manifestations of alpha-synucleinopathy in vivo.
Accumulation of toxic alpha-synuclein oligomer within endoplasmic reticulum occurs in alpha-synucleinopathy in vivo. Conn, K. Brain Res. Cooper, A. Coppola-Segovia, V. Corona Velazquez, A. So many roads: the multifaceted regulation of autophagy induction. Cortes, C.
TFEB dysregulation as a driver of autophagy dysfunction in neurodegenerative disease: molecular mechanisms, cellular processes, and emerging therapeutic opportunities.
Costa, C. Cells Coune, P. Cold Spring Harb. Credle, J. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Cuervo, A. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Dagda, R. Autophagy 4, — De Miranda, B. Decressac, M.
TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Deegan, S. Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress.
Dehay, B. Deng, C. Inhibition of 6-hydroxydopamine-induced endoplasmic reticulum stress by sulforaphane through the activation of Nrf2 nuclear translocation. Duplan, E. ER-stress-associated functional link between Parkin and DJ-1 via a transcriptional cascade involving the tumor suppressor p53 and the spliced X-box binding protein XBP Cell Sci.
Egawa, N. The endoplasmic reticulum stress sensor, ATF6alpha, protects against neurotoxin-induced dopaminergic neuronal death. Esteves, A. Fiesel, F. Patho- physiological relevance of PINK1-dependent ubiquitin phosphorylation. Disease relevance of phosphorylated ubiquitin p-SUb.
Autophagy 11, — Gade, P. Regulation of the death-associated protein kinase 1 expression and autophagy via ATF6 requires apoptosis signal-regulating kinase 1. Gambardella, G. GADD34 is a modulator of autophagy during starvation.
Gao, S. Gardner, B. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response.
Ghemrawi, R. Endoplasmic reticulum stress and unfolded protein response in neurodegenerative diseases. Ghribi, O. Giordano, S. Bioenergetic adaptation in response to autophagy regulators during rotenone exposure.
Gitler, A. Gonzalez-Polo, R. Silencing DJ-1 reveals its contribution in paraquat-induced autophagy. Gorbatyuk, M. Glucose regulated protein 78 diminishes alpha-synuclein neurotoxicity in a rat model of Parkinson disease. Grassi, D. Gupta, S. Salubrinal attenuates nitric oxide mediated PERK:IRE1alpha: ATF-6 signaling and DNA damage in neuronal cells.
Harding, H. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Cell 6, — CrossRef Full Text Google Scholar. Haze, K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress.
Cell 10, — Heman-Ackah, S. Hetz, C. The unfolded protein response and cell fate control. Cell 69, — Hillary, R. A lifetime of stress: ATF6 in development and homeostasis. Ho, D. LRRK2 impairs autophagy by mediating phosphorylation of leucyl-tRNA synthetase.
Cell Biochem. Ho, P. Autophagy 16, — Holtz, W. Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons.
Hoozemans, J. Hou, X. Age- and disease-dependent increase of the mitophagy marker phospho-ubiquitin in normal aging and Lewy body disease. Autophagy 14, — Imai, Y. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell , — Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity.
Iurlaro, R. Cell death induced by endoplasmic reticulum stress. FEBS J. Iwata, A. alpha-Synuclein affects the MAPK pathway and accelerates cell death. Jiang, P. ER stress response plays an important role in aggregation of alpha-synuclein.
Jiang, T. Neuroimmune Pharmacol. Jiao, F. Jovanovic-Tucovic, M. Kalia, L. Lancet , — Kalogeropulou, A. Kalvakolanu, D. IFNG and autophagy: a critical role for the ER-stress mediator ATF6 in controlling bacterial infections.
Autophagy 8, — Karabiyik, C. Essays Biochem. Kitada, T. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature , — Korennykh, A. Structural basis of the unfolded protein response. Cell Dev. Kouroku, Y. Lamark, T.
NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 8, — Ledesma, M. Astrocytic but not neuronal increased expression and redistribution of parkin during unfolded protein stress.
Lee, D. PARK7 modulates autophagic proteolysis through binding to the N-terminally arginylated form of the molecular chaperone HSPA5. Lee, J. Acta Neuropathol. Lehtonen, S. Levine, B. Biological functions of autophagy genes: a disease perspective.
Cell , 11— Li, D. Additionally, PERK activates its downstream signaling eIF2a-ATF4-CHOP pathway and NF-κB which initiates inflammation and apoptosis.
ERS leads to dissociation of TRAF from TRAF2-procaspase 12 complex, which is located on the ER membrane, leading to activation of caspase The ATF6 pathway also activates NF-κB, further intensifying the expression of inflammatory genes, which secrete more cytokines.
PRR induced-inflammation is beneficial in mounting an immune response against microbes. However, profound inflammation in the absence of infection is pathologic which is supported by the clinical use of antibodies against inflammatory cytokines to treat diseases such as intestinal bowel disease, rheumatoid arthritis, and multiple sclerosis.
As discussed above, UPR signaling and autophagy are intertwined with inflammation Figure 2. Therefore, UPR has been linked with several inflammatory diseases and some of which has been reviewed here below.
Figure 2. Crosstalk between ER stress, autophagy and inflammation. The arms of UPR activating inflammation also intersect with pathways regulating autophagy. The possible points of intersection are shown in the illustration. Intestinal epithelial cells IECs are constantly exposed to microbiota, metabolites and toxins which force them to produce large amounts of cytokines and various other proteins resulting in ER-stress.
Although, UPR helps in resolving ER-stress, continued ER-stress and disruptions in the UPR mechanism can result in chronic inflammation. Therefore, it is no surprise that studies have associated UPR dysregulation with Crohn's disease CD and ulcerativecolitis UC , two major types of IBDs 47 , IBD is also one of the first polygenic disease to be genetically linked to UPR components Intestinal inflammation is primarily linked to IRE1-XBP1 arm of the UPR pathway because mice deleted of the IRE1 gene in mouse intestinal epithelium are more susceptible to dextran sulfate sodium DSS -induced colitis Similarly, mice deficient in XBP-1 in the intestine develop spontaneous intestinal inflammation and immune infiltration resembling IBD The barrier between microbial flora of the gut and IECs is maintained by the secretion of mucin2 MUC2.
UPR and intestinal inflammation has been also linked in humans. For instance, anterior gradient 2 AGR2 encoding a protein-disulfide-isomerase which enables protein folding and orosomucoid-like 3 ORMDL3 , which regulates ER calcium have been shown to induce UPR 53 — Furthermore, genome-wide association studies GWAS have mapped the XBP-1 gene locus as an IBD susceptibility region 57 , As described before, UPR interacts with autophagy pathways at multiple levels.
UPR induces autophagy and reciprocally, autophagy may limit UPR by reducing ER-stress Interestingly, a core autophagy effector protein ATG16L is associated with IBD in humans. Consistently, mice deficient in ATG16L1 in IECs develop Crohn's like disease 60 — Furthermore, deletion of ATG16L1 and XBP1 in IECs results in more severe IBD suggesting that autophagy and UPR synergizes in regulating intestinal inflammation UPR is also associated with the pathogenesis of chronic obstructive pulmonary disease COPD.
External stimulants such as cigarette smoke induces ROS production which disturbs the redox environment thus preventing proper protein folding by modulating the protein-disulfide-isomerase PDI Dysregulation of protein folding in lung and bronchial epithelial cells induces UPR 64 , Furthermore, oxidative damage of proteins caused by cigarette smoke leads to impaired degradation of misfolded, non-functional proteins triggering UPR Cigarette smoke induced-UPR is characterized by PERK-eIF2a-mediated CHOP induction 64 , 66 — Impaired autophagy has been linked to cigarette smoke induced inflammation.
On the contrary, activating autophagy using mTOR inhibitor rapamycin results in increased apoptosis and inflammation Interestingly, another form of autophagy known as chaperone-mediated autophagy CMA , which is LAMP2A facilitated selective degradation of proteins containing Lys-Phe-Gln-Arg-Gln KFERQ in the lysosomes mitigates cigarette smoke induced UPR and apoptosis MS is an autoimmune disorder in which the T-cells target myelin sheath ER-stress induced UPR is found to be a hallmark of MS It is proposed that autophagy-induced cell death could be a possible mechanism by which UPR resolves ER-stress.
Hence, autophagy is elevated in MS-lesions resulting in demyelination and neuro-inflammation. PERK and CHOP activation has been found to be consistent with upregulation of BAX and BCL2 in experimental autoimmune encephalomyelitis EAE.
However, the molecular mechanisms integrating UPR, autophagy and inflammation has not been completely understood 3. Parkinson's Disease PD is a neurodegenerative disease and numerous evidences suggest that inflammation exacerbates the disease Reports have also linked the role of ER-stress in the pathogenesis of PD using neurotoxic models of PD.
Interestingly, depletion of CHOP protects dopaminergic neurons against hydroxydopamine 6-OHDA indicating the involvement of ER-stress in PD Similarly, silencing XBP1 another UPR arm results in chronic ER stress and dopaminergic neuron degeneration Parkin an E3 ubiquitin ligase implicated in Parkinson's disease is a key regulator of mitochondria-specific autophagy mitophagy.
Interestingly, ATF4 upregulates parkin by directly binding to the promoter region upon ER stress Although, studies addressing the cross talk between ER stress, autophagy and inflammation in PD are limited, UPR can co-regulate inflammation and autophagy as discussed above.
Intriguingly, ER stress that is implicated in inflammation and autophagy has been currently coupled with the pathophysiological aspects of the cardiovascular system CVS Upregulation of UPR is observed in cardiac hypertrophy and heart failure. Inflammation and ER stress within the CVS are connected through various regulators such as NF-κB, JNK, spliced XBP-1 and ROS 80 — As discussed in earlier sections of this review, UPR activation leads to recruitment of TRAF2 by IRE1 which interacts with JNK and IκB resulting in the activation of downstream inflammatory signaling and cytokine production.
Additionally, IRE1 auto-phosphorylates and splices its downstream XBP-1 which stimulates the production of inflammatory cytokines by enhancing Toll-like receptor TLR signaling 32 , ATF6 activation also results in transcriptional activation of inflammatory proteins like C-reactive protein CRP which fosters the expression of monocyte chemoattractant protein-1 MCP-1 and contributes to inflammation 84 , Furthermore, ATF6 phosphorylates AKT and activates NF-κB which stimulates the expression of various cytokines Similarly, PERK also triggers NF-κB-induced cytokine signaling by activating IκB It is well-established that ER stress is also implicated in atherosclerosis where UPR activation is observed in macrophage-derived and smooth muscle cell SMC -derived foam cells 79 , The plaque deposition in the arterial walls triggers infiltration of macrophages and neutrophils leading to production of IL-1 and IL-6 Additionally, ROS is induced resulting in UPR activation which can further enhance inflammation and tissue damage Inflammation induced mitochondrial damage and ROS can in turn induce autophagy A weak association between autophagy and plaque formation has been reported based on the expression of autophagy markers 91 , However, whether autophagy is beneficial or detrimental in atherosclerosis is poorly understood.
Traditionally, engagement of PRRs with PAMPs has been considered the primary trigger for inflammation.
However, changes in intracellular functions causing cellular stress have been lately recognized to play a key role in inflammation associated with pathologies. Thus, ER stress-induced inflammation has been implicated in several inflammatory diseases.
Although response to ER-stress UPR aids in mitigating ER-stress, UPR pathways also promote inflammation and diseases such as diabetes, obesity, IBD, inflammatory lung disorders, cardiovascular diseases and cancer.
Moreover, UPR pathways are interlinked with other cellular-stress response mechanisms such as autophagy which can potentially mitigate inflammation and disease progression. Conversely, activation of cellular homeostasis mechanisms such as autophagy can be an impediment to treat diseases such as cancer.
However, UPR induced inflammation and autophagy vary between diseases and is cell type dependent. Although, inflammation and autophagy have been reported during ER-stress, it is correlative.
Therefore, molecular mechanisms that integrate UPR, autophagy and inflammation need to be elucidated which is crucial for therapeutic targeting. SC: prepared the manuscript draft and figures. All authors contributed to the article and approved the submitted version. Research in the laboratory of NR was supported by funds from the Center for Cancer Biology, University of South Australia and Neurosurgical Research Foundation, Adelaide, Australia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers.
Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher. Hotamisligil GS. Inflammation and metabolic disorders. doi: PubMed Abstract CrossRef Full Text Google Scholar.
Schröder M, Kaufman RJ. ER stress and the unfolded protein response. Mut Res Fund Mol Mech Mutagen. Andhavarapu S, Mubariz F, Arvas M, Bever C Jr, Makar TK.
Interplay between ER stress and autophagy: a possible mechanism in multiple sclerosis pathology. Exp Mol Pathol. Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response.
Matsuzawa-Ishimoto Y, Hwang S, Cadwell K. Autophagy and inflammation. Ann Rev Immunol. Lin JH, Walter P, Yen TB. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol Mech Dis. Schönthal AH. Endoplasmic reticulum stress: its role in disease and novel prospects for therapy.
Ghemrawi R, Battaglia-Hsu S-F, Arnold C. Endoplasmic reticulum stress in metabolic disorders. Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response.
Nat Rev Mol Cell Biol. Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol.
Chipurupalli S, Kannan E, Tergaonkar V, D'Andrea R, Robinson N. Hypoxia induced ER stress response as an adaptive mechanism in cancer. Int J Mol Sci. Kopp MC, Larburu N, Durairaj V, Adams CJ, Ali MM.
UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor. Nat Struct Mol Biol. Adams CJ, Kopp MC, Larburu N, Nowak PR, Ali MM.
Structure and molecular mechanism of ER stress signaling by the unfolded protein response signal activator IRE1. Front Mol Biosci. Parmar VM, Schröder M. Sensing Endoplasmic Reticulum Stress Self and Nonself. New York, NY: Springer Google Scholar.
Hetz C, Zhang K, Kaufman RJ. Mechanisms, regulation and functions of the unfolded protein response. Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Tepedelen BE, Kirmizibayrak PB. Endoplasmic Reticulum-Associated Degradation ERAD.
In Endoplasmic Reticulum. London: Intechopen. Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. Tsai YC, Weissman AM. The unfolded protein response, degradation from the endoplasmic reticulum, and cancer. Genes Cancer.
Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Verfaillie T, Salazar M, Velasco G, Agostinis P.
Linking ER stress to autophagy: potential implications for cancer therapy. Int J Cell Biol. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy.
Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response.
Nat Cell Biol. Corazzari M, Rapino F, Ciccosanti F, Giglio P, Antonioli M, Conti B, et al. Oncogenic BRAF induces chronic ER stress condition resulting in increased basal autophagy and apoptotic resistance of cutaneous melanoma.
Cell Death Different. B'chir W, Maurin A-C, Carraro V, Averous J, Jousse C, Muranishi Y, et al. Nucl Acids Res. B'chir W, Chaveroux C, Carraro V, Averous J, Maurin A-C, Jousse C, et al.
Dual role for CHOP in the crosstalk between autophagy and apoptosis to determine cell fate in response to amino acid deprivation. Cell Sign. Chaudhari N, Talwar P, Parimisetty A, Lefebvre d'Hellencourt C, Ravanan P.
A molecular web: endoplasmic reticulum stress, inflammation, oxidative stress. Front Cell Neurosci. Garg AD, Kaczmarek A, Krysko O, Vandenabeele P, Krysko DV, Agostinis P. ER stress-induced inflammation: does it aid or impede disease progression? Trends Mol Med. Pahl HL. Chen ZJ. Ubiquitin signalling in the NF-κB pathway.
Riaz TA, Junjappa RP, Handigund M, Ferdous J, Kim H-R, Chae H-J, et al. Role of endoplasmic reticulum stress sensor ire1α in cellular physiology, calcium, ROS signaling, and metaflammation.
Liu Y-P, Zeng L, Tian A, Bomkamp A, Rivera D, et al. Endoplasmic reticulum stress regulates the innate immunity critical transcription factor IRF3. J Immunol. Smith JA. Regulation of cytokine production by the unfolded protein response; implications for infection and autoimmunity.
Front Immunol. West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Martinon F, Chen X, Lee A-H, Glimcher LH.
TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. Zeng L, Liu Y-P, Sha H, Chen H, Qi L, et al. XBP-1 couples endoplasmic reticulum stress to augmented IFN-β induction via a cis-acting enhancer in macrophages.
Iwasaki Y, Suganami T, Hachiya R, Shirakawa I, Kim-Saijo M, Tanaka M, et al. Activating transcription factor 4 links metabolic stress to interleukin-6 expression in macrophages. Goodall JC, Wu C, Zhang Y, McNeill L, Ellis L, Saudek V, et al.
Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL expression. Proc Natl Acad Sci USA. Lerner AG, Upton J. IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress.
Cell Metab. Oslowski CM, Hara T, O'Sullivan-Murphy B, Kanekura K, Lu S, Hara M, et al. Thioredoxin-interacting protein mediates ER stress-induced β cell death through initiation of the inflammasome. Dong D, Fu N, Yang P. MiR downregulation by high glucose stabilizes thioredoxin-interacting protein and removes thioredoxin inhibition on ASK1 leading to apoptosis.
Toxicol Sci. Keestra-Gounder AM, Byndloss MX, Seyffert N, Young BM, Chávez-Arroyo A, Tsai AY, et al. NOD1 and NOD2 signalling links ER stress with inflammation.
Shenderov K, Riteau N, Yip R, Mayer-Barber KD, Oland S, Hieny S, et al. Cutting edge: endoplasmic reticulum stress licenses macrophages to produce mature IL-1β in response to TLR4 stimulation through a caspase-8—and TRIF-dependent pathway. Smith JA, Turner MJ, DeLay ML, Klenk EI, Sowders DP, Colbert RA.
Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-β induction via X-box binding protein 1. Eur J Immunol. Hu F, Yu X, Wang H, Zuo D, Guo C, Yi H, et al. ER stress and its regulator X-box-binding protein-1 enhance polyIC-induced innate immune response in dendritic cells.
Kaser A, Lee A. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease.
McGovern DP, Gardet A, Törkvist L, Goyette P, Essers J, Taylor KD, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Gen. Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, et al.
Increased sensitivity to dextran sodium sulfate colitis in IRE1β-deficient mice. Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, et al. IRE1 signaling affects cell fate during the unfolded protein response.
Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. Wenzel UA, Jonstrand C, Hansson GC, Wick MJ.
PLoS ONE. Zheng W, Rosenstiel P, Huse K, Sina C, Valentonyte R, Mah N, et al.
The endoplasmic strews ER is not strses responsible strfss protein synthesis and folding but also Antioxidant enzymes a critical role in sensing cellular stress and maintaining cellular homeostasis. Autophagy and ER stress sensing the sttess of unfolded proteins due to perturbation in protein Autophagy and ER stress or anx, specific intracellular signaling Type diabetes symptoms are Autophagy and ER stress, which are collectively termed as unfolded protein response UPR. UPR expands the capacity of the protein folding machinery, decreases protein synthesis and enhances ER-associated protein degradation ERAD which degrades misfolded proteins through the proteasomes. More recent evidences suggest that UPR also amplifies cytokines-mediated inflammatory responses leading to pathogenesis of inflammatory diseases. UPR signaling also activates autophagy; a lysosome-dependent degradative pathwaythat has an extended capacity to degrade misfolded proteins and damaged ER. Thus, activation of autophagy limits inflammatory response and provides cyto-protection by attenuating ER-stress. Here we review the mechanisms that couple UPR, autophagy and cytokine-induced inflammation that can facilitate the development of novel therapeutic strategies to mitigate cellular stress and inflammation associated with various pathologies. Division of Endocrinology and Metabolism, Department of Internal Anv, Seoul National University Ahd, Seoul, Korea. Download PDF. This was ans by NRF grants R1A2C and R1A2C funded by the Ministry of Stfess and ICT, Republic of Autophavy. Skip Navigation Autophaby to contents Search Autophagy and ER stress Current Current issue Ahead-of print Browse All issues Article by category Autophagy and ER stress by topic Article by Category Best paper of Autopgagy year Most Auto;hagy Most Digestive aid for digestive enzyme support Funded srress Diabetes Metab J Search Author index Collections Autophaty in DMJ Fact sheets in DMJ COVID in DMJ For contributors For Authors Instructions to authors Article processing charge e-submission For Reviewers Instructions for reviewers How to become a reviewer Best reviewers For Readers Readership Subscription Permission guidelines About Aims and scope About the journal Editorial board Management team Best practice Metrics Contact us Editorial policy Research and publication ethics Peer review policy Copyright and open access policy Article sharing author self-archiving policy Archiving policy Data sharing policy Preprint policy Advertising policy E-Submission. mobile menu button. Author information Article notes Copyright and License information Division of Endocrinology and Metabolism, Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea Corresponding author: Hye Seung Jung Division of Endocrinology and Metabolism, Department of Internal Medicine, Seoul National University Hospital, Daehak-ro, Jongno-gu, SeoulKorea E-mail address: jungjhs gmail. ABSTRACT Pancreatic beta cell homeostasis is crucial for the synthesis and secretion of insulin; disruption of homeostasis causes diabetes, and is a treatment target.
Ich wollte dieses Thema nicht entwickeln.
entschuldigen Sie, ich habe nachgedacht und hat die Mitteilung gelöscht
Im Vertrauen gesagt, ich empfehle Ihnen, in google.com zu suchen