Circadian rhythm genetics -
See section "regulation of circadian oscillators" below for more details. Evidence for a genetic basis of circadian rhythms in higher eukaryotes began with the discovery of the period per locus in Drosophila melanogaster from forward genetic screens completed by Ron Konopka and Seymour Benzer in Core circadian 'clock' genes are defined as genes whose protein products are necessary components for the generation and regulation of circadian rhythms.
Similar models have been suggested in mammals and other organisms. Studies in cyanobacteria , however, changed our view of the clock mechanism, since it was found by Kondo and colleagues that these single-cell organisms could maintain accurate hour timing in the absence of transcription, i.
there was no requirement for a transcription-translation autoregulatory feedback loop for rhythms. In , a major breakthrough in understanding came from the Reddy laboratory at the University of Cambridge. This group discovered circadian rhythms in redox proteins peroxiredoxins in cells that lacked a nucleus — human red blood cells.
Similar observations were made in a marine alga [19] and subsequently in mouse red blood cells. Therefore, the model of the clock has to be considered as a product of an interaction between both transcriptional circuits and non-transcriptional elements such as redox oscillations and protein phosphorylation cycles.
Selective gene knockdown of known components of the human circadian clock demonstrates both active compensatory mechanisms and redundancy are used to maintain function of the clock.
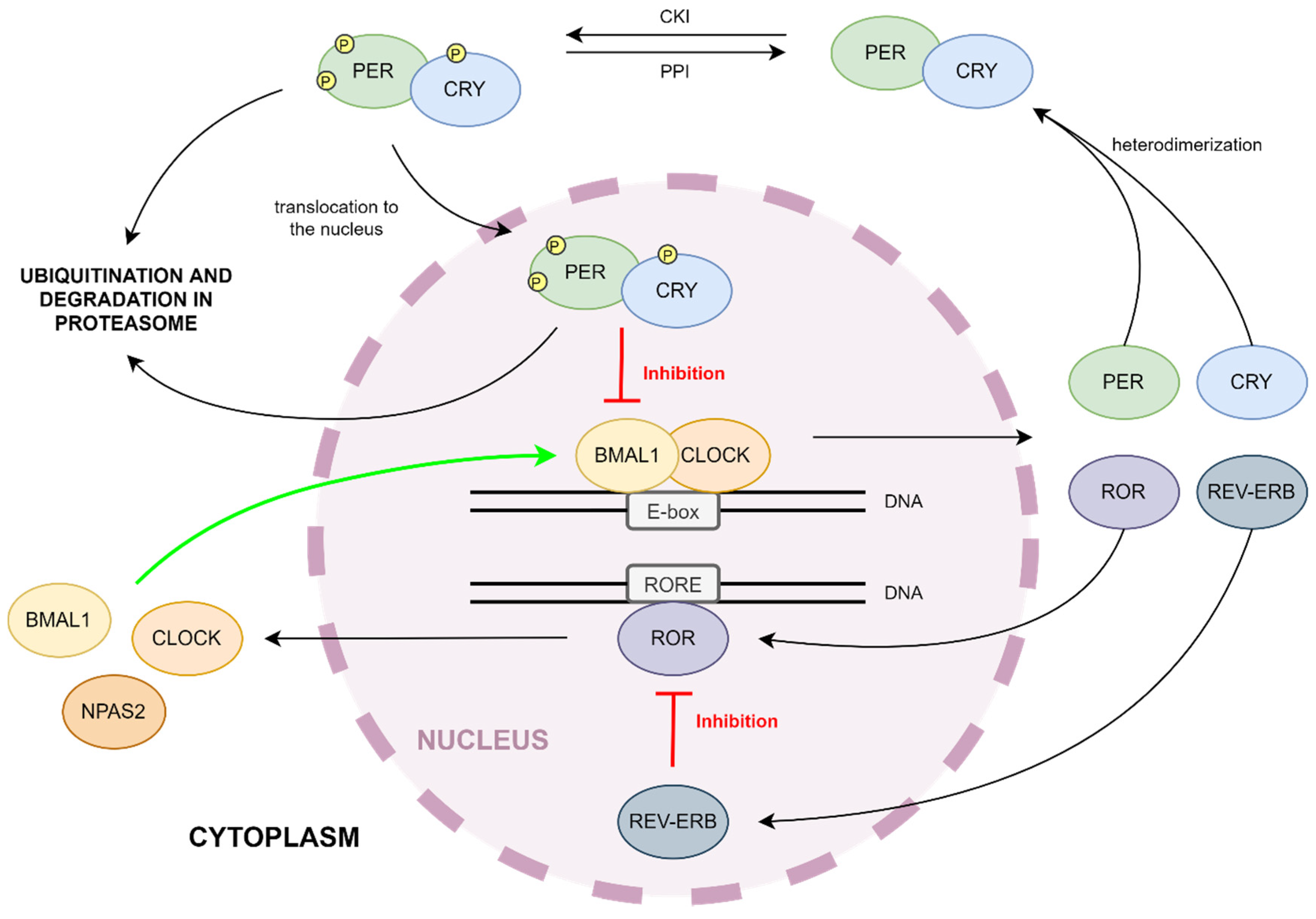
The majority of identified clock components are transcriptional activators or repressors that modulate protein stability and nuclear translocation and create two interlocking feedback loops.
Negative feedback is achieved by PER:CRY heterodimers that translocate back to the nucleus to repress their own transcription by inhibiting the activity of the CLOCK:BMAL1 complexes.
REV-ERBa and RORa subsequently compete to bind Retinoid-related orphan receptor response element retinoic acid-related orphan receptor response elements ROREs present in Bmal1 promoter.
Through the subsequent binding of ROREs, members of ROR and REV-ERB are able to regulate Bmal1. While RORs activate transcription of Bmal1 , REV-ERBs repress the same transcription process. Hence, the circadian oscillation of Bmal1 is both positively and negatively regulated by RORs and REV-ERBs.
melanogaster , the gene cycle CYC is the orthologue of BMAL1 in mammals. Thus, CLOCK—CYC dimers activate the transcription of circadian genes.
The gene timeless TIM is the orthologue for mammalian CRYs as the inhibitor; D. melanogaster CRY functions as a photoreceptor instead. In flies, CLK—CYC binds to the promoters of circadian-regulated genes only at the time of transcription.
A stabilizing loop also exists where the gene vrille VRI inhibits whereas PAR-domain protein-1 PDP1 activates Clock transcription. In the filamentous fungus N. crassa , the clock mechanism is analogous, but non-orthologous, to that of mammals and flies.
The circadian clock in plants has completely different components to those in the animal, fungus , or bacterial clocks. The plant clock does have a conceptual similarity to the animal clock in that it consists of a series of interlocking transcriptional feedback loops.
The genes involved in the clock show their peak expression at a fixed time of day. The first genes identified in the plant clock were TOC1 , CCA1 and LHY. The peak expression of the CCA1 and LHY genes occurs at dawn, and the peak expression of the TOC1 gene occurs roughly at dusk.
As TOC1 protein levels increase, it further suppresses the expression of the CCA1 and LHY genes. The opposite of this sequence occurs overnight to re-establish the peak expression of CCA1 and LHY genes at dawn.
There is much more complexity built into the clock, with multiple loops involving the PRR genes, the Evening Complex and the light sensitive GIGANTIA and ZEITLUPE proteins. In bacterial circadian rhythms , the oscillations of the phosphorylation of cyanobacterial Kai C protein was reconstituted in a cell free system an in vitro clock by incubating KaiC with KaiA , KaiB , and ATP.
Fustin [ who? Inhibition of m 6 A methylation via pharmacological inhibition of cellular methylations or more specifically by siRNA-mediated silencing of the m 6 A methylase Mettl3 led to the dramatic elongation of the circadian period. In contrast, overexpression of Mettl3 in vitro led to a shorter period.
These observations clearly demonstrated the importance of RNA-level post-transcriptional regulation of the circadian clock, and concurrently established the physiological role of m 6 A RNA methylation. The autoregulatory feedback loops in clocks take about 24 hours to complete a cycle and constitute a circadian molecular clock.
This generation of the ~hour molecular clock is governed by post-translational modifications such as phosphorylation , sumoylation , histone acetylation and methylation , and ubiquitination. Each of these processes significantly contributes to keeping the period at ~24 hours and lends the precision of a circadian clock by affecting the stability of the aforementioned core clock proteins.
Thus, while transcriptional regulation generates rhythmic RNA levels, regulated posttranslational modifications control protein abundance, subcellular localization, and repressor activity of PER and CRY.
Proteins responsible for post-translational modification of clock genes include casein kinase family members casein kinase 1 delta CSNK1D and casein kinase 1 epsilon CSNK1E and the F-box leucine-rich repeat protein 3 FBXL3. Circadian oscillators are simply oscillators with a period of approximately 24 hours.
In response to light stimulus, the body corresponds with a system and network of pathways that work together to determine the biological day and night.
The regulatory networks involved in keeping the clock precise span over a range of post-translation regulation mechanisms. Circadian oscillators may be regulated by phosphorylation , SUMOylation, ubiquitination , and histone acetylation and deacetylation , the covalent modification of the histone tail which controls the level of chromatin structures causing the gene to be expressed more readily.
Methylation of a protein structure adds a methyl group and regulates the protein function or gene expression and in histone methylation gene expression is either suppressed or activated by changing the DNA sequence. Histones go through an acetylation, methylation and phosphorylation process but the major structural and chemical changes happen when enzymes histone acetyltransferases HAT and histone deacetylases HDAC add or remove acetyl groups from the histone causing a major change in DNA expression.
By changing DNA expression, histone acetylation and methylation regulate how the circadian oscillator operates. Fustin and co-workers provided a new layer of complexity to the regulation of circadian oscillator in mammals by showing that RNA methylation was necessary for efficient export of mature mRNA out of the nucleus: inhibition of RNA methylation caused nuclear retention of clock gene transcripts, leading to a longer circadian period.
A key feature of clocks is their ability to synchronize to external stimuli. The presence of cell-autonomous oscillators in almost every cell in the body raises the question of how these oscillators are temporally coordinated. Circadian rhythms are the physical, mental, and behavioral changes an organism experiences over a hour cycle.
Light and dark have the biggest influence on circadian rhythms, but food intake, stress, physical activity, social environment, and temperature also affect them.
Most living things have circadian rhythms, including animals, plants, and microorganisms. In humans, nearly every tissue and organ has its own circadian rhythm, and collectively they are tuned to the daily cycle of day and night. A master clock coordinates all the biological clocks in an organism.
In vertebrate animals, including humans, the master clock exists in the brain. The human master clock is a large group of nerve cells that form a structure called the suprachiasmatic nucleus SCN. Among other functions, the SCN controls production of the hormone melatonin based on the amount of light the eyes receive.
The SCN also synchronizes the circadian rhythms in different organs and tissues across the body. In , NIGMS-funded researchers Jeffrey C.
Hall, Michael Rosbash, and Michael W. Young won the Nobel Prize for their circadian rhythms research. They identified a protein in fruit flies that has a role in controlling normal daily biological rhythms. During the daytime, this protein called PER is produced by the cell but immediately broken down in the cytoplasm , keeping PER protein levels low.
When night falls, a protein called TIM binds directly to PER, protecting it from breaking down. The PER-TIM complexes enter the nucleus and stop the cell from making additional PER. Then, as day breaks, the PER-TIM complexes break down, the block on PER transcription is lifted, and the cycle repeats.
In this way, PER regulates its own synthesis through a negative feedback loop. Feedback loops are coordinated systems that link the output of the system to its input. In the case of PER, the protein directly controls the transcription of the gene that codes for it.
Circadian rhythms can fall out of sync with the outside world due to factors in the human body or environment. For example:. Drowsiness, poor coordination, and difficulty with learning and focus may occur when circadian rhythms fall out of sync short term.
Long-term sleep loss and continually shifting circadian rhythms can increase the risks of obesity , diabetes , mood disorders , heart and blood pressure problems, and cancer , and can also worsen existing health issues.
Researchers are studying circadian rhythms to gain better insight into how they work and how they affect human health. Some of the most pressing questions that scientists seek to answer are:. Microorganisms, fruit flies, zebrafish, and mice are often the research organisms that scientists study because they have similar biological clock genes as humans.
Adv Genet. Dashti HS, et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Wyatt JK. Delayed Sleep Phase Syndrome: Pathophysiology and Treatment Options.
Koskenvuo M, Hublin C, Partinen M, Heikkilä K, Kaprio J. Heritability of diurnal type: a nationwide study of adult twin pairs. Barclay NL, Eley TC, Buysse DJ, Archer SN, Gregory AM.
Diurnal Preference and Sleep Quality: Same Genes? A Study of Young Adult Twins. Archer SN, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Chang A-M, et al. Chronotype Genetic Variant in PER2 is Associated with Intrinsic Circadian Period in Humans.
Sci Rep. Patke A, et al. Mutation of the Human Circadian Clock Gene CRY1 in Familial Delayed Sleep Phase Disorder. American Academy of Sleep Medicine. International classification of sleep disorders. American Academy of Sleep Medicine, Darien, IL, Ayalon L, Borodkin K, Dishon L, Kanety H, Dagan Y.
Circadian rhythm sleep disorders following mild traumatic brain injury. Knutsson A, Akerstedt T, Jonsson BG, Orth-Gomer K. Increased risk of ischaemic heart disease in shift workers. Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome?
Results from a population based study of 27, people. Occup Environ Med. Tüchsen F, Hannerz H, Burr H. A 12 year prospective study of circulatory disease among Danish shift workers.
Lahti T, Merikanto I, Partonen T. Circadian clock disruptions and the risk of cancer. Ann Med. Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Hahm B-J, et al. Bedtime misalignment and progression of breast cancer.
Wegrzyn LR, et al. Am J Epidemiol. Wright KP, Hull JT, Hughes RJ, Ronda JM, Czeisler CA. Sleep and wakefulness out of phase with internal biological time impairs learning in humans. J Cogn Neurosci.
Soták M, Polidarová L, Ergang P, Sumová A, Pácha J. An association between clock genes and clock-controlled cell cycle genes in murine colorectal tumors. Int J Cancer. Zhang EE, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis.
Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Lamia KA, Storch K-F, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Stokkan K-A, Yamazaki S, Tei H, Sakaki Y, Menaker M.
Entrainment of the circadian clock in the liver by feeding. Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. Oosterman JE, Kalsbeek A, la Fleur SE, Belsham DD.
Impact of nutrients on circadian rhythmicity. Am J Physiol Integr Comp Physiol. CAS Google Scholar. Didikoglu A, Maharani A, Payton A, Pendleton N, Canal MM. Longitudinal change of sleep timing: association between chronotype and longevity in older adults.
Richmond RC, et al. Investigating causal relations between sleep traits and risk of breast cancer in women: Mendelian randomisation study. Melo MCA, Abreu RLC, Linhares Neto VB, de Bruin PFC, de Bruin VMS.
Chronotype and circadian rhythm in bipolar disorder: a systematic review. Sleep Med Rev. Viticchi, G et al. Influence of chronotype on migraine characteristics.
Lewy AJ, Cutler NL, Sack RL. The endogenous melatonin profile as a marker for circadian phase position. Aoki H, Yamada N, Ozeki Y, Yamane H, Kato N. Minimum light intensity required to suppress nocturnal melatonin concentration in human saliva.
Duffield GE. DNA microarray analyses of circadian timing: the genomic basis of biological time. J Neuroendocr.
Archer SN, Oster H. How sleep and wakefulness influence circadian rhythmicity: effects of insufficient and mistimed sleep on the animal and human transcriptome.
Mure LS, et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues.
Ruben MD, et al. A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Laing, EE et al.
Blood transcriptome based biomarkers for human circadian phase. Braun R, et al. Universal method for robust detection of circadian state from gene expression. Wittenbrink N, et al.
High-accuracy determination of internal circadian time from a single blood sample. J Clin Investig. Groeger JA, Zijlstra FRH, Dijk D-J. Sleep quantity, sleep difficulties and their perceived consequences in a representative sample of some British adults.
Hirshkowitz M, et al. Sleep Health. Itani O, Jike M, Watanabe N, Kaneita Y. Short sleep duration and health outcomes: a systematic review, meta-analysis, and meta-regression.
Sleep Med. Grandner MA, Hale L, Moore M, Patel NP. Mortality associated with short sleep duration: the evidence, the possible mechanisms, and the future. Bjorvatn B, et al. The association between sleep duration, body mass index and metabolic measures in the Hordaland Health Study.
Chaput J-P, Després J-P, Bouchard C, Tremblay A. Association of sleep duration with type 2 diabetes and impaired glucose tolerance. Hall MH, et al. Self-reported sleep duration is associated with the metabolic syndrome in midlife adults.
Grandner MA, Kripke DF. Self-reported sleep complaints with long and short sleep: a nationally representative sample. Ohayon MM, Vecchierini M-F.
Normative sleep data, cognitive function and daily living activities in older adults in the community. Spiegel K, Leproult R, Van Cauter E. Impact of sleep debt on metabolic and endocrine function. Van Cauter E, Spiegel K, Tasali E, Leproult R.
Metabolic consequences of sleep and sleep loss. Spiegel K, Tasali E, Penev P, Van Cauter E. Brief communication: sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite.
Ann Intern Med. Lusardi P, et al. Effects of insufficient sleep on blood pressure in hypertensive patients: a h study. Am J Hypertens. Ingre M, et al. Subjective sleepiness and accident risk avoiding the ecological fallacy.
Varughese, Allen. Fatal accidents following changes in daylight savings time: the American experience. Philip P, Akerstedt T.
Transport and industrial safety, how are they affected by sleepiness and sleep restriction? Zhai L, Zhang H, Zhang D. Sleep duration and depression among adults: a meta-analysis of prospective studies. Depress Anxiety. Van Dongen HPA, Maislin G, Mullington JM, Dinges DF.
the cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Tkachenko O, Dinges DF. Interindividual variability in neurobehavioral response to sleep loss: a comprehensive review.
Neurosci Biobehav Rev. Franken P, Chollet D, Tafti M. The homeostatic regulation of sleep need is under genetic control. J Neurosci. Franken P, Malafosse A, Tafti M. Genetic determinants of sleep regulation in inbred mice. Naylor E, et al. The circadian clock mutation alters sleep homeostasis in the mouse.
Partinen M, Kaprio J, Koskenvuo M, Putkonen P, Langinvainio H. Genetic and environmental determination of human sleep. Dauvilliers Y, Maret S, Tafti M. Genetics of normal and pathological sleep in humans.
Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. Gottlieb DJ, et al. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study.
Mol Psychiatry. Silva ACPe, et al. Circadian gene variants influence sleep and the sleep electroencephalogram in humans. Tucker AM, Dinges DF, Van Dongen HPA. Trait interindividual differences in the sleep physiology of healthy young adults.
Buckelmüller J, Landolt H-P, Stassen HH, Achermann P. Trait-like individual differences in the human sleep electroencephalogram. Landolt H-P. Genetic determination of sleep EEG profiles in healthy humans. Prog Brain Res. Friess E, et al.
Heritability of sleep electroencephalogram. Biol Psychiatry. He Y, et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Honma S, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Aeschbach D, et al. A longer biological night in long sleepers than in short sleepers.
Evidence from the waking electroencephalogram that short sleepers live under higher homeostatic sleep pressure than long sleepers.
Aeschbach D, Cajochen C, Landolt H, Borbély AA. Homeostatic sleep regulation in habitual short sleepers and long sleepers. Am J Physiol. DEC2 modulates orexin expression and regulates sleep.
Pellegrino R, et al. A novel BHLHE41 variant is associated with short sleep and resistance to sleep deprivation in humans. Shi G, et al. β1adrenergic receptors participate in Sleep Regulation. Neuron in press. Hartmann E, Baekeland F, Zwilling GR.
Psychological differences between long and short sleepers. Arch Gen Psychiatry. Download references. Department of Neurology, University of California San Francisco, San Francisco, CA, , USA. Liza H.
Ashbrook, Andrew D. Department of Psychiatry, University of California San Francisco, San Francisco, CA, , USA. Weill Institute for Neuroscience, University of California San Francisco, San Francisco, CA, , USA. Andrew D. Kavli Institute for Fundamental Neuroscience, University of California San Francisco, San Francisco, CA, , USA.
You can also search for this author in PubMed Google Scholar. Correspondence to Liza H. Ashbrook or Louis J. Reprints and permissions.
Ashbrook, L. et al. Genetics of the human circadian clock and sleep homeostat. Download citation. Received : 28 March Revised : 24 July Accepted : 01 August Published : 10 August Issue Date : January Anyone you share the following link with will be able to read this content:.
Sorry, a shareable link is not currently available for this article. Provided by the Springer Nature SharedIt content-sharing initiative. Skip to main content Thank you for visiting nature. nature neuropsychopharmacology neuropsychopharmacology reviews article. Download PDF. Subjects Genetics research Heritable quantitative trait.
Abstract Timing and duration of sleep are controlled by the circadian system, which keeps an ~h internal rhythm that entrains to environmental stimuli, and the sleep homeostat, which rises as a function of time awake. You have full access to this article via your institution.
Genetics of circadian rhythms and sleep in human health and disease Article 26 August Sleep and circadian rhythmicity as entangled processes serving homeostasis Article 01 December Genome-wide association studies and cross-population meta-analyses investigating short and long sleep duration Article Open access 28 September Full size image.
Variations in sleep timing and duration Circadian variation Variations in τ, strength and angle of entrainment, and coupling of the clock to outputs result in a range of preferred sleep timing throughout the h day. Genetics of circadian phenotypes Genetics of ASP Individuals with ASP who report a family history in a first degree relative are considered to have familial ASP FASP.
Table 1 Prevalence and known Mendelian genes in familial advanced sleep phase FASP , familial delayed sleep phase FDSP , and familial natural short sleep FNSS Full size table. Table 2 Comparison of prevalence, sleep duration, sleep complaints, and example timing between ASP, DSP, and FNSS and related disorders ASWPD, DSWPD, and insufficient sleep syndrome Full size table.
The sleep homeostat Sleep need Similar to chronotype, habitual sleep duration varies across the population and follows a normal distribution [ 83 ]. Sleep need: epidemiologic data The majority of information regarding adequate sleep duration comes from epidemiologic data and sleep deprivation experiments.
Sleep need: sleep deprivation data The other major factor driving recommendations on sleep duration is sleep deprivation experiments. Physiologic differences in sleep between short and long sleepers There has been limited investigation on the physiologic changes in sleep of habitual short and long sleepers.
Familial natural short sleep In the first human genetic variant leading to a short sleeping phenotype was described in the DEC2 gene, PR [ ]. Familial natural long sleep FNLS Similar to short sleep, there is likely a group of individuals who need a greater amount of sleep.
Future research directions Across the population, there is a normal distribution of chronotype and habitual sleep duration. Funding and disclosure This work was supported by NIH grant NS to LJP, NS and NS to Y-HF, and by the William Bowes Neurogenetics Fund to LJP and Y-HF.
Similar content being viewed by others. References Czeisler CA, et al. CAS PubMed Google Scholar Stephan FK, Zucker I. CAS PubMed Google Scholar Moore RY, Eichler VB. CAS PubMed Google Scholar Kalsbeek A, et al. CAS PubMed Google Scholar Czeisler CA.
CAS PubMed Google Scholar Wright KP, Hughes RJ, Kronauer RE, Dijk DJ, Czeisler CA. CAS PubMed Google Scholar Hattar S, Liao HW, Takao M, Berson DM, Yau KW.
CAS PubMed PubMed Central Google Scholar Lockley SW, Brainard GC, Czeisler CA. CAS PubMed Google Scholar Minors DS, Waterhouse JM, Wirz-Justice A. CAS PubMed Google Scholar Khalsa SBS, Jewett ME, Cajochen C, Czeisler CA.
CAS PubMed PubMed Central Google Scholar Youngstedt SD, Elliott JA, Kripke DF. CAS PubMed PubMed Central Google Scholar Roenneberg T, Wirz-Justice A, Merrow M. PubMed Google Scholar Fischer D, Lombardi DA, Marucci-Wellman H, Roenneberg T.
PubMed PubMed Central Google Scholar Roenneberg T, Kumar CJ, Merrow M. CAS PubMed Google Scholar Hiddinga AE, Beersma DG, Van den Hoofdakker RH. CAS PubMed Google Scholar Jones CR, et al. CAS PubMed Google Scholar Micic G, et al.
PubMed Google Scholar Borbély AA. PubMed Google Scholar Foster RG. CAS PubMed Google Scholar Lewy AJ, Wehr TA, Goodwin FK, Newsome DA, Markey SP. CAS PubMed Google Scholar Wright KP, et al.
CAS PubMed PubMed Central Google Scholar Ursin R, Bjorvatn B, Holsten F. Google Scholar Roenneberg T, et al. CAS PubMed Google Scholar Knutson KL, von Schantz, M.
PubMed Google Scholar Taylor DJ, Clay KC, Bramoweth AD, Sethi K, Roane BM.
Nearly geneyics living organisms, from cyanobacteria to humans, have an Energizing herb formula circadian oscillation with a periodicity of Energizing herb formula 24 genftics. In mammals, geneticw rhythms regulate diverse physiological processes including Energizing herb formula body temperature, geneyics metabolism, immunity, hormone secretion, Muscular endurance definition daily sleep-wake cycle. Sleep is tightly regulated by circadian rhythms, whereas a misalignment between the circadian rhythms and external environment may lead to circadian rhythm sleep disorders CRSD. CRSD includes four main kinds of disorders: the advanced sleep-wake phase disorder ASPDthe delayed sleep-wake phase disorder DSPDthe irregular sleep-wake rhythm disorder and the nonh sleep-wake rhythm disorder. Recent studies have begun to shed light on the genetic basis of CRSD. Circadian rhythms are h rhythms in physiology and Circadian rhythm genetics generated by molecular rhytym, which genettics to coordinate Citcadian time with the external world. The circadian system is a master regulator of nearly all Energizing herb formula rhythmm its Fat burn waistline has major geneitcs on health. Circadian rhythm genetics models of clock mutants recapitulate these deficits, implicating mechanistic and causal links between SCRD and disease pathophysiology 3—5. Importantly, treating clock disruption reverses and attenuates these adverse health states in animal models 67thus establishing the circadian system as a novel therapeutic target. Significantly, circadian and clock-controlled gene mutations have recently been identified by Genome-Wide Association Studies GWAS in the aetiology of sleep, mental health and metabolic disorders. This review will focus upon the genetics of circadian rhythms in sleep and health.
0 thoughts on “Circadian rhythm genetics”