
Video
MBS Lehninger Chapter 22 Biosynthesis of Amino AcidsAmino acid biosynthesis -
Amino acids are organic compounds that consist of alpha carbon in the center, hydrogen H , amino -NH2 , carboxyl -COOH , and specific R side chain groups.
One linear chain of amino acids is called a polypeptide, and one or more polypeptides make up a protein. There are 20 major types of amino acids found in proteins, of which the differences are the side chains R groups that contain various chemical structures.
This R group gives each amino acid and, finally, each protein-specific characteristic. These features include size, shape, hydrophilicity, hydrophobicity, interactions, polarity, and pH level.
Each of these characteristics is crucial for the stability of the proteins in the human body and environment. As building blocks of proteins, amino acids are essential for multiple biological processes, including cell growth, division, and metabolic signaling pathways.
However, the dysregulation of pathways involved in amino acid biogenesis and catabolism have been characterized in multiple inborn metabolic disorders such as phenylketonuria PKU , alkaptonuria, and maple syrup urine disease MSUD.
Gly, Ala, Val, Leu, Met, and Ile have aliphatic R groups, and Phe, Tyr, and Trp have aromatic R groups. The aliphatic or aromatic group makes the amino acids hydrophobic also called "water fear" or group with no tendency to be close to aqueous solutions.
The globular proteins will opportunistically bury the hydrophobic side chains inside the protein interior by folding into a three-dimensional shape in aqueous solutions.
The amino acids with polar uncharged R groups are serine S, Ser, HO-CH2-CH NH2 -COOH , threonine T, Thr, CH3-CH OH -CH NH2 -COOH , cysteine C, Cys, HS-CH2-CH NH2 -COOH , proline P, Pro, NH- CH2 3-CH-COOH , asparagine N, Asn, H2N-CO-CH2-CH NH2 -COOH and glutamine Q, Gln, H2N-CO- CH2 2-CH NH2 -COOH.
The side chains of these amino acids possess functional spectrum groups. Most have one or more atoms, such as oxygen, nitrogen, or sulfur, with electron pairs, allowing hydrogen bonding to water or other molecules.
Also, aspartate D, Asp, HOOC-CH2-CH NH2 -COOH and glutamate E, Glu, HOOC- CH2 2-CH NH2 -COOH are amino acids with negatively charged R groups. The amino acids are linked with their neighbors in a specific order by covalent bonds, also known as peptide bonds.
These particular bonds are the amide linkages that form when the amino group reacts with the carboxylate carbon connecting two amino acids. The free amino group at one end of the polypeptide is typically called the amino-terminal or N-terminal.
In contrast, the open carboxyl group at the other end is labeled as the carboxyl-terminal or C-terminal. Protein sequences are written or read from the N-to-C terminal direction. The chains of the amino acids or progression of the amino acids distinguish exquisitely one protein from another.
The organism's DNA is specific in coding a particular sequence of amino acids. Each protein consists of one or more polypeptide chains. Proteins are polymers of 50 or more amino acids, while peptides are shorter amino acid polymers.
A protease, which is also known as peptidase or proteinase, is an enzyme that catalyzes proteolysis. This phenomenon is constituted by the breakdown of proteins into smaller polypeptides and, eventually, single amino acids. Proteases cleave the peptide bonds within proteins by hydrolysis, which is a chemical reaction where the water breaks bonds.
Acids, alkalis, or enzymes may be employed to determine protein hydrolysis. The general functions of amino acids include the involvement in protein synthesis, biosynthetic products, and metabolic energy.
Essentially, there is a crucial difference between positive and negative nitrogen balance, which is critical for understanding amino acid metabolism. In a positive balance, the nitrogen consumed is more considerable than the nitrogen excreted, while in a negative balance, the nitrogen consumed is less than the nitrogen excreted.
A positive balance denotes net protein synthesis. It occurs when the organism is recovering from starvation, growth, and pregnancy. In contrast, a negative balance entails mobilization of the amino acids, tissue necrosis, or a poor-quality condition of the human body as a consequence of 3rd-degree burns or significant surgical operations.
The amino acids subdivide into essential and non-essential. There are amino acids that need to be obtained directly diet and amino acids that can be synthesized by the organism. The amino acids the human body cannot produce are called essential amino acids, which contain His, Ile, Leu, Lys, Met, Val, Phe, Thr, and Trp.
The human body gets these nine essential amino acids from food or nutritional supplement. In specific medical conditions or at different ages, the other amino acids may be conditionally essential for the human body.
Glutamate is a non-essential amino acid that can be synthesized from alpha-ketoglutaric acid in the Krebs or citric acid cycle. In the brain and spinal cord, glutamate is synthesized from glutamine as part of the glutamate-glutamine cycle by the enzyme glutaminase.
Glutamate cannot cross the blood-brain barrier unaided and serves as a metabolic precursor for the neurotransmitter gamma-aminobutyric acid GABA via the action of glutamate decarboxylase. Methionine is converted to S-adenosylmethionine SAM by methionine adenosyltransferase.
Loss of methionine has correlated with an accumulation of hydrogen peroxide H2O2 in hair follicles, a decrease in tyrosinase effectiveness, and a gradual loss of the natural hair color.
GSH is an antioxidant found in animals, plants, fungi, bacteria, and archaea. Promoting antioxidant-mediated cell defense and redox regulation is critical in protecting cells against dopamine-induced nigral cell loss by oxidative binding metabolites.
These amino acids are cysteine, carnitine, taurine, lecithin, and phosphatidylcholine. Also, methionine is medium in the biosynthesis of additional phospholipids. Improper transformation of methionine can lead to atherosclerosis due to the accumulation of homocysteine. Moreover, this amino acid is essential to reversing the damaging methylation of the glucocorticoid receptor gene caused by repeated stress exposures, with implications for depression.
Glycine is considered to be not essential to the human diet. The body can synthesize this amino acid from the amino acid serine.
However, the metabolic capacity for glycine biosynthesis does not satisfy the need for collagen synthesis in several organisms. In the liver of some of them at the vertebrate level, glycine synthesis is catalyzed by glycine synthase, which is also known as glycine cleavage enzyme.
Glycine is integral to the creation of alpha-helices in secondary protein structure, and, mainly, it is the most copious amino acid in collagen harboring triple-helices. Glycine is also an inhibitory neurotransmitter. The interference of its release within the central nervous system spinal cord can induce spastic paralysis due to uninhibited muscle contraction.
Amino acids are synthesized through different pathways. Cys is synthesized from Met, while Tyr synthesis can occur using Phe, considering that the amino acid precursors can be available in the body. The amino acid Arg, which arises from the urea cycle, is considered "semi-essential" because the synthetic capacity of the human body is limited.
Non-essential amino acids need their precursors, which must be available in the organism. Specifically, Ala and Gly's amino acids need pyruvate to be synthesized, while aspartic acid and Asn rely on oxaloacetic acid OAA.
Thus, six essential amino acids and three non-essential are integrated from oxaloacetate and pyruvate. The transamination from Glu is vital in forming Asp and Ala from OAA and pyruvate.
Asp is crucial in synthesizing Asn, Met, Lys, and Thr. OAA is critical because no Asp would form without it. The alpha-ketoglutaric acid or 2-oxoglutaric acid is one of two ketone derivatives of glutaric acid. Its anion, alpha-ketoglutarate alpha-KG , also known as 2-oxoglutarate, is a biological compound of paramount importance.
It is the keto acid produced by the deamination of Glu and is an intermediate compound in the urea or Krebs cycle. The amino acids glutamic acid and Gln arise from alpha-KG.
Finally, the amino acid Pro derives from Glu, while Ser is from 3-phosphoglyceric acid 3PG. The 3PG is the conjugate acid of glycerate 3-phosphate.
It is a biochemically significant metabolic intermediate in glycolysis and the Calvin cycle. In the Calvin cycle or photosynthetic carbon reduction PCR cycle of photosynthesis, 3PG is vital. It is the product of the spontaneous scission of an unstable 6-carbon intermediate formed upon CO fixation.
Thus, glycerate 3-phosphate is a precursor for Ser, which, in turn, can create Cys and Gly through the homocysteine cycle. Therefore, Pro arises from Glu, while Ser is from 3PG.
In the transamination reaction, an amino acid Ala or Asp exchanges its amine group for the oxy group in alpha-KG. The products are Glu and pyruvate or OAA from Ala or Asp, accordingly. Different proteases can degrade proteins into many small peptides or amino acids by hydrolyzing their peptide bonds.
The unused amino acids may degrade further to join several metabolic processes. At first, the amino acids deaminate to their metabolic intermediates. This process is helpful for the excretion of an excessive amount of nitrogen.
Subsequently, they can transform into the remaining carbon skeleton. In particular, this deamination process contains two steps.
The first part uses deamination. In this step, the aminotransferase catalyzes the -NH2 group of the amino acid to alpha-KG. After that, it produces Glu and a novel alpha-keto acid of the specific amino acid.
The Glu -NH2 group could then be transferred to OAA to form alpha-KG and Asp. This trans-amination series only degrade the primary amino acid, while the -NH2 group nitrogen does not exclude. Then, it produces ammonia and alpha-KG. In the evaluation of the biochemistry of the amino acids, seven metabolic intermediates of the aminoacidic degradation platform are paramount.
They include acetyl-CoA, pyruvate, alpha-KG, acetoacetate, fumarate, succinyl-CoA, and OAA. In the most updated classification, Leu, Ile, Thr, and Lys degrade to acetyl-CoA, while Cys, Ala, Thr, Gly, Trp, and Ser degrade to pyruvate. Glu, Arg, His, Pro, and Gln degrade to alpha-KG, while Lys, Leu, Trp, Tyr, and Phe break down to acetoacetate.
Finally, Tyr, Phe, and Asp degrade to fumarate, Val, Met, and Ile break down to succinyl-CoA, and Asp and, of course, Asn degrade to OAA. Isoleucine is an essential nutrient because it is unsynthesized in the body.
This amino acid is both a glucogenic and ketogenic amino acid. In microorganisms and plants, it is synthesized via several steps beginning with pyruvate and alpha-ketobutyrate. The enzymes involved in this biosynthesis include acetolactate synthase, acetohydroxy acid isomeroreductase, dihydroxy acid dehydratase, and valine aminotransferase.
In clinical practice, plasma or urine amino acids undergo testing to evaluate patients with possible inborn metabolism problems. They can also assess many diseases, such as liver diseases, endocrine disorders, muscular diseases, neurological disorders, neoplastic diseases, renal failure, burns, and nutritional disturbances.
Both high-performance liquid chromatography HPLC and gas chromatography GC have been used to quantitatively identify the plasma or urine amino acids in clinical settings.
Amino acid disorders are identifiable at any age; most of them become evident during infancy or early childhood. Many inborn amino metabolism diseases occur in infancy or childhood. These disorders may include cystinuria, histidinemia, phenylketonuria PKU , methyl-malonyl CoA mutase deficiency MCM deficiency , albinism, and tyrosinemia.
Other amino acid disorders may be encountered later in life, including homocystinuria, alkaptonuria, maple syrup urine disease MSUD , and cystathioninuria. These disorders lead to clinical symptoms or signs of the specific amino acid disorder, which results in the deficiency or accumulation of one or more amino acids in the body's biological fluids, such as plasma or urine.
The deficiency of Phe hydroxylase causes PKU. Yusa, K. A hyperactive piggyBac transposase for mammalian applications. Chatterjee, A. Efficient viral delivery system for unnatural amino acid mutagenesis in mammalian cells. Tanaka-Azevedo, A.
Thrombin Inhibitors from Different Animals. Koh, C. Molecular diversity of anticoagulants from haematophagous animals. Kazimírová, M. Tick salivary compounds: their role in modulation of host defences and pathogen transmission. Hsieh, Y. Total synthesis of homogeneous variants of hirudin P6: a post-translationally modified anti-thrombotic leech-derived protein.
Corral-Rodríguez, M. Leech-derived thrombin inhibitors: from structures to mechanisms to clinical applications. Watson, E. Mosquito-Derived Anophelin Sulfoproteins Are Potent Antithrombotics. ACS Cent. Rapid assembly and profiling of an anticoagulant sulfoprotein library.
Nakajima, C. A Novel Gene Encoding a Thrombin Inhibitory Protein in a cDNA Library from Haemaphysalis longicornis Salivary Gland. Young, D. Playing with the Molecules of Life. Xiao, H. At the Interface of Chemical and Biological Synthesis: An Expanded Genetic Code. Cold Spring Harb.
a Iannuzzelli, J. Expanded toolbox for directing the biosynthesis of macrocyclic peptides in bacterial cells. Owens, A. MOrPH-PhD: An Integrated Phage Display Platform for the Discovery of Functional Genetically Encoded Peptide Macrocycles. Huang, Y. Therapeutic applications of genetic code expansion.
Chen, C. Genetic-code-expanded cell-based therapy for treating diabetes in mice. Zhang, S. A general strategy to red-shift green fluorescent protein-based biosensors.
Chen, Z. Genetically Encoded Fluorescent Probe for the Selective Detection of Peroxynitrite. Guo, J. Genetic Code Expansion Through Quadruplet Codon Decoding.
Download references. We thank Dr. Xiao Laboratory members for insightful comments. This work was supported by the Cancer Prevention Research Institute of Texas CPRIT RR to H. Welch Foundation C to H. Dunn Foundation Collaborative Research Award to H.
and D. Bullard Welch Chair at Rice University Grant C to P. Department of Chemistry, Rice University, Main Street, Houston, TX, , USA. Yuda Chen, Mengxi Zhang, Yu Hu, Kuan-Lin Wu, Anna Chung, Shichao Wang, Zeru Tian, Yixian Wang, Peter G.
Center for Theoretical Biological Physics, Rice University, Main Street, Houston, TX, , USA. Department of Biosciences, Rice University, Main Street, Houston, TX, , USA. Department of Physics, Rice University, Main Street, Houston, TX, , USA. Department of Bioengineering, Rice University, Main Street, Houston, TX, , USA.
You can also search for this author in PubMed Google Scholar. designed the project. and Y. constructed plasmids. and K. performed the SSN analysis. conducted structure prediction and docking experiments.
expressed and purified proteins. carried out confocal microscopy. All other experiments were performed by Y. and H. wrote the paper. Correspondence to Han Xiao. Nature Communications thanks Nediljko Budisa and the other, anonymous, reviewer s for their contribution to the peer review of this work.
Open Access This article is licensed under a Creative Commons Attribution 4. Reprints and permissions. Unleashing the potential of noncanonical amino acid biosynthesis to create cells with precision tyrosine sulfation. Nat Commun 13 , Download citation.
Received : 21 March Accepted : 01 September Published : 16 September Anyone you share the following link with will be able to read this content:. Sorry, a shareable link is not currently available for this article. Provided by the Springer Nature SharedIt content-sharing initiative.
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily. Skip to main content Thank you for visiting nature. nature nature communications articles article.
Download PDF. Subjects Protein engineering Proteins Synthetic biology. Abstract Despite the great promise of genetic code expansion technology to modulate structures and functions of proteins, external addition of ncAAs is required in most cases and it often limits the utility of genetic code expansion technology, especially to noncanonical amino acids ncAAs with poor membrane internalization.
Introduction With the rare exceptions of pyrrolysine and selenocysteine, a standard set of 20 amino acid building blocks, containing a limited number of functional groups, is used by almost all organisms for the biosynthesis of proteins. Full size image. Molecular basis of Nn SULT1C1 action in the sulfation of tyrosine To explore the origin of the unique tyrosine specificity of Nn SULT1C1 among all the sulfotransferases tested, we analyzed the phylogenetic relationships of the enzymes.
Discussion In this research, we have generated completely autonomous bacterial and mammalian cells endowed with machinery for both sTyr biosynthesis and site-specific incorporation into proteins. Predicting the structure of Nn SULT1C1 by AlphaFold The structure of Nn SULT1C1 was predicted by AlphaFold2 using GitHub AlphaFold code 2.
Protein-ligand docking The protein-ligand docking process was performed by Glide v8. Thrombin activity assay N- p -Tosyl -GPR- p NA acetate Cayman Chemicals was used as a chromogenic substrate to test the amidolytic activity of human α-thrombin Haematologic Technologies.
Statistics and reproducibility All statictics analysis were performed using GraphPad Prism. Reporting summary Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability All data generated in this study are included in the paper and supplementary information. References Wang, L. Article PubMed CAS Google Scholar Ambrogelly, A. Article CAS PubMed Google Scholar Liu, C.
Article CAS PubMed Google Scholar Chin, J. Article CAS PubMed Google Scholar Dien, V. Article CAS PubMed PubMed Central Google Scholar Chin, J. Article ADS CAS PubMed Google Scholar Agostini, F.
Article CAS Google Scholar Manandhar, M. Article CAS PubMed Google Scholar Wang, L. Article ADS CAS PubMed Google Scholar Furter, R. Article CAS PubMed PubMed Central Google Scholar Giese, C.
Article CAS PubMed Google Scholar Zhang, M. Article CAS PubMed PubMed Central Google Scholar Luo, X. Article CAS PubMed PubMed Central Google Scholar Hoppmann, C. Article CAS PubMed PubMed Central Google Scholar Bundy, B. Article CAS PubMed Google Scholar Ko, W.
Article CAS PubMed Google Scholar Burkovski, A. Article CAS PubMed Google Scholar Palacín, M. Article PubMed Google Scholar Mehl, R.
Article CAS PubMed Google Scholar Chen, Y. Article CAS Google Scholar Rogerson, D. Article CAS PubMed PubMed Central Google Scholar Chen, Y. Article CAS PubMed PubMed Central Google Scholar Völler, J. Article PubMed CAS Google Scholar Wang, Y.
Article CAS Google Scholar Exner, M. Article PubMed PubMed Central CAS Google Scholar Ludeman, J. Article CAS PubMed PubMed Central Google Scholar Farzan, M. Article CAS PubMed Google Scholar Choe, H. Article CAS PubMed Google Scholar Thompson, R.
Article CAS PubMed Google Scholar Somers, W. Article CAS PubMed Google Scholar Westmuckett, A. Article ADS CAS PubMed PubMed Central Google Scholar Lee, S.
Article ADS CAS PubMed Google Scholar Li, X. Article CAS PubMed PubMed Central Google Scholar Liu, C. Article ADS CAS PubMed PubMed Central Google Scholar Liu, C. Article CAS PubMed Google Scholar Schwessinger, B. Article CAS Google Scholar Yang, Y.
Article PubMed PubMed Central CAS Google Scholar Gamage, N. Article CAS PubMed Google Scholar Allali-Hassani, A. Article PubMed PubMed Central CAS Google Scholar Suiko, M. Article CAS PubMed PubMed Central Google Scholar Jendresen, C.
Article Google Scholar Chen, Y. Article CAS PubMed PubMed Central Google Scholar Copp, J. Article CAS PubMed Google Scholar Gerlt, J. Article CAS PubMed PubMed Central Google Scholar Blanchard, R. Article CAS PubMed Google Scholar Schlee, D. Article Google Scholar Varin, L.
Article CAS PubMed Google Scholar Hirschmann, F. Article ADS CAS PubMed PubMed Central Google Scholar Tunyasuvunakool, K. Article ADS CAS PubMed PubMed Central Google Scholar Jin, S. Article CAS PubMed Google Scholar Jin, S.
Article PubMed PubMed Central Google Scholar Berger, I. Article ADS CAS PubMed PubMed Central Google Scholar Lu, J.
Article CAS PubMed Google Scholar Bidwell, L. Article CAS PubMed Google Scholar Friesner, R. Article CAS PubMed Google Scholar Banks, J. Article CAS PubMed PubMed Central Google Scholar Zhu, S.
Article ADS PubMed PubMed Central CAS Google Scholar Wei, T. Article PubMed PubMed Central CAS Google Scholar Chu, L.
Article PubMed CAS Google Scholar Neuwald, A. Article CAS PubMed PubMed Central Google Scholar Spiegelberg, B. Article CAS PubMed Google Scholar Lu, L. Article CAS PubMed Google Scholar Dajani, R.
Article CAS PubMed Google Scholar Moore, K. Article ADS CAS PubMed PubMed Central Google Scholar Seibert, C. Article CAS PubMed Google Scholar He, X. Article ADS CAS PubMed PubMed Central Google Scholar Italia, J.
Article CAS PubMed PubMed Central Google Scholar Yusa, K. Article ADS CAS PubMed PubMed Central Google Scholar Chatterjee, A. Article ADS CAS PubMed PubMed Central Google Scholar Tanaka-Azevedo, A. Article CAS PubMed Google Scholar Kazimírová, M. Article CAS PubMed Google Scholar Corral-Rodríguez, M.
Article PubMed CAS Google Scholar Watson, E. Article CAS PubMed PubMed Central Google Scholar Watson, E. Article ADS CAS PubMed PubMed Central Google Scholar Nakajima, C.
Article CAS PubMed Google Scholar Young, D. Article CAS PubMed PubMed Central Google Scholar Xiao, H. Article CAS PubMed PubMed Central Google Scholar Owens, A.
Article CAS PubMed PubMed Central Google Scholar Huang, Y. Article PubMed PubMed Central Google Scholar Chen, C. Article CAS PubMed Google Scholar Zhang, S.
Article CAS PubMed PubMed Central Google Scholar Chen, Z. Article CAS PubMed Google Scholar Guo, J. Acknowledgements We thank Dr. Author information Authors and Affiliations Department of Chemistry, Rice University, Main Street, Houston, TX, , USA Yuda Chen, Mengxi Zhang, Yu Hu, Kuan-Lin Wu, Anna Chung, Shichao Wang, Zeru Tian, Yixian Wang, Peter G.
Wolynes Department of Biosciences, Rice University, Main Street, Houston, TX, , USA Shikai Jin, Peter G. Wolynes Department of Bioengineering, Rice University, Main Street, Houston, TX, , USA Han Xiao Authors Yuda Chen View author publications. View author publications.
Ethics declarations Competing interests The authors declare no competing interests. Peer review Peer review information Nature Communications thanks Nediljko Budisa and the other, anonymous, reviewer s for their contribution to the peer review of this work.
Supplementary information. Supplementary Information. Reporting Summary. Description of Additional Supplementary Files. Supplementary Data 1. Source data Source Data. Rights and permissions Open Access This article is licensed under a Creative Commons Attribution 4.
About this article. Cite this article Chen, Y. Copy to clipboard. This article is cited by Enhanced tyrosine sulfation is associated with chronic kidney disease-related atherosclerosis Daopeng Dai Zhengbin Zhu Jinzhou Zhu BMC Biology A platform for distributed production of synthetic nitrated proteins in live bacteria Neil D.
Butler Sabyasachi Sen Aditya M. For isoleucine, 2-oxobutanoate is converted to 2-acetohydroxybutanoate, then to 2,3-dihydroxymethylpentanoate, then to 3-methyloxopentanoate, and finally to isoleucine.
The final steps in the biosynthesis of both BCAAs can be performed by native CHO catabolic enzymes Bcat1 and Bcat2 Hefzi et al. The final pMTIV construct comprises metC , itaE , ilvN, ilvB , ilvC, and ilvD organized as a single open reading frame ORF with a 2A sequence variant lying between each protein coding region Figure 2B , and driven by a single strong spleen focus-forming virus SFFV promoter.
A Three enzymatic steps encoded by E. coli genes ilvN regulatory subunit, acetolactate synthase , ilvB catalytic subunit, acetolactate synthase , ilvC ketol-acid reductoisomerase , and ilvD dihydroxy-acid dehydratase are required for valine biosynthesis in Chinese hamster ovary CHO -K1 cells.
B Schematic of pMTIV construct after genomic integration and RNA-seq read coverage showing successful incorporation and active transcription. C Microscopy images of CHO-K1 cells with integrated pCtrl or pMTIV constructs in complete FK medium after 2 days or valine-free FK medium after 6 days.
Scale bar represents µm. D Growth curve of CHO-K1 cells with pCtrl or pMTIV in complete FK medium Figure 2—source data 1. Day-0 indicates number of seeded cells. Error bars represent data from three replicates. E Growth curve of CHO-K1 cells with pCtrl or pMTIV in valine-free FK medium Figure 2—source data 1.
Raw cell count data for pMTIV valine-free and complete FK medium tests. To test the biosynthetic capacity of pMTIV, we first introduced the construct into CHO cells.
Flp-In integration was used to stably insert either pMTIV, or a control vector pCtrl into the CHO genome. Successful generation of each cell line was confirmed by PCR amplification of junction regions formed during vector integration Figure 2—figure supplement 2A-B.
RNA-seq of cells containing the pMTIV construct confirmed transcription of the entire ORF Figure 2B. Western blotting of pMTIV cells using antibodies against the P2A peptide yielded bands at the expected masses of P2A-tagged proteins, confirming the production of separate distinct enzymes Figure 2—figure supplement 2C.
In reconstituted methionine-free, threonine-free, or isoleucine-free FK medium supplemented with dialyzed FBS to reduce FBS-derived AA content Figure 2—figure supplement 3 , cells containing the pMTIV construct did not show viability over 7 days, similar to cells containing the pCtrl control vector Figure 2—figure supplement 4.
In striking contrast, however, cells containing the integrated pMTIV showed relatively healthy cell morphology and viability in valine-free FK medium Figure 2C , whereas cells containing pCtrl exhibited substantial loss of viability over 6 days.
In complete FK medium, cells carrying the integrated pMTIV construct showed no growth defects compared to control cells Figure 2D. When cultured in valine-free FK medium over multiple passages with medium changes every 2 days, pMTIV cell proliferation was substantially reduced by the 3rd passage.
We hypothesized that frequent passaging might over-dilute the medium and prevent sufficient accumulation of biosynthesized valine necessary for continued proliferation as has been demonstrated for certain non-essential metabolites which become essential when cells are cultured at low cell densities Eagle and Piez, While use of pMTIV-conditioned medium improved the survival of cells harboring the pathway, it did not completely rescue valine auxotrophy in control cells, which exhibited substantial loss of cell viability over 8 days Figure 2—figure supplement 5A.
As a control, we generated pCtrl-conditioned valine-free FK medium using the same medium conditioning regimen, which failed to enable cells to grow to the same degree as that of pMTIV-conditioned medium, suggesting that the benefit conferred by medium conditioning is valine-specific Figure 2—figure supplement 5B.
Using this regimen, we were able to culture pMTIV cells for 9 passages without addition of exogenous valine Figure 2F. The doubling time was inconsistent across the 49 days of experimentation with cells exhibiting a mean doubling time of 5.
Despite the slowed growth seen in later passages, cells exhibited healthy morphology and continued viability at day, suggesting that the cells could have been passaged even further. The pIV construct similarly supported cell growth in valine-free FK medium, and exhibited similar growth dynamics to the pMTIV construct in complete medium Figure 2—figure supplement 6.
To confirm endogenous biosynthesis of valine, we cultured pCtrl and pMTIV cells in RPMI medium containing 13 C 6 -glucose in the place of its 12 C equivalent together with 13 C 3 -sodium pyruvate spiked in at 2 mM over three passages Figure 3—figure supplement 1A.
High-resolution MS1 of MTIV cell lysates revealed a peak at The resulting fragmentation patterns for each peak Figure 3B matched theoretical expectations for each isotopic version of valine Figure 3—figure supplement 1B. Taken together, this demonstrates that pMTIV cells are capable of biosynthesizing valine from core metabolites glucose and pyruvate, thereby proving successful metazoan biosynthesis of valine.
Over the course of 3 passages in heavy valine-free medium, the non-essential amino acid alanine, which is absent from RPMI medium and synthesized from pyruvate, was found to be Assuming similar turnover rates for alanine and valine within the CHO proteome, we expected to see similar percentages of 13 C-labeled valine.
However, just For pMTIV cells cultured in heavy but valine-replete medium, just 6. Together with the observed slow proliferation of pMTIV cells in valine-free medium, our data suggests that valine complementation is sufficient but sub-optimal for cell growth.
MS2 fragmentation patterns for each of these metabolites matched expectations Figure 3—source data 1. C RNA-seq dendrogram of pCtrl cells and pMTIV cells grown on complete FK medium or starved of valine for 4 hr or 48 hr. D Principal Component Analysis PCA space depiction of pCtrl cells and pMTIV cells grown on complete FK medium, or starved of valine for 4 hr or 48 hr.
We performed RNA-seq to profile the transcriptional responses of cells containing pMTIV or pCtrl in complete harvested at 0 hr and valine-free FK medium harvested at 4 hr and 48 hr, respectively Figure 3C , Figure 3—figure supplement 2A.
The transcriptional impact of pathway integration is modest Figure 3D. Only 51 transcripts were differentially expressed between pCtrl and pMTIV cells grown in complete medium, and the fold changes between conditions were small Figure 3E , Figure 3—figure supplement 2B.
While some gene ontology GO functional categories were enriched Figure 3—figure supplement 2C , they did not suggest dramatic cellular stress. Rather, these transcriptional changes may reflect cellular response to BCAA dysregulation due to altered valine levels Zhenyukh et al.
In contrast, comparison of 48 hr valine-starved pCtrl and pMTIV cells yielded ~ differentially expressed genes. Transcriptomes of pMTIV cells in valine-free medium more closely resembled cells grown on complete medium than did pCtrl cells in valine-free medium Figure 3D , Figure 3—figure supplement 3A.
Differentially expressed genes between pCtrl and pMTIV cells showed enrichment for hundreds of GO categories, including clear signatures of cellular stress such as autophagy, changes to endoplasmic reticulum trafficking, and ribosome regulation Figure 3—figure supplement 3B.
Most of the differentially regulated genes between pCtrl cells in complete medium, and those same cells starved of valine for 48 hr were also differentially expressed when comparing pCtrl and pMTIV cells in valine-free medium Figure 3E , supporting the hypothesis that most of the observed transcriptional changes represent broad but partial rescue of the cellular response to starvation.
We also examined the integrated stress response ISR and mTOR signaling pathways, both of which are known to modulate cellular responses to starvation Pakos-Zebrucka et al. We observed no clear signatures of mTOR activation Figure 3—figure supplement 4A , although a number of individual genes related to the mTOR pathway were significantly differentially expressed compared to pCtrl cells valine-starved for 48 hr Figure 3—figure supplement 4B.
A manually curated list of ISR genes showed signals of ISR gene activation, but showed few differences between pCtrl and pMTIV cells at 48 hr of starvation Figure 3—figure supplement 4C.
pMTIV cells grown for 5 passages over 29 days on conditioned valine-free FK medium were more similar to pMTIV cells starved for 48 hr than to pCtrl cells starved for 48 hr Figure 3—figure supplement 5A.
To improve rescue of the valine starvation phenotype, we looked for valine biosynthetic pathway intermediates in our metabolomics data that might suggest that the pathway was bottlenecked at any stage.
While no signal could be detected for pyruvate, 2-acetolactate or 2-oxoisovalerate, a signal was detected for pathway intermediate 13 C 5 -2,3-dihydroxy-isovalerate, which was specific to pMTIV cells cultured in both complete and in valine-free medium Figure 3—figure supplement 1F.
To determine whether the downstream pathway gene, ilvD , which encodes the dihydroxy-acid dehydratase enzyme, might constitute a bottleneck, we generated a lentivirus encoding a puromycin resistance cassette in addition to ilvD under control of a viral MMLV promoter Figure 4A.
Both pCtrl and pMTIV cells were infected and integrants were selected for on puromycin, resulting in a population-averaged integration count of 5.
This resulted in a 0. B ilvD qPCR on gDNA and cDNA from each cell line Figure 4—source data 1. Fold change levels were relativized to pMTIV. cDNA was reverse transcribed using oligo dT primers from RNA templates collected from each cell line.
Error bars show SD of three technical replicates. Error bars represent data from two replicates. We also reduced the isoleucine content of this isotopically heavy valine-free RPMI medium to match the isoleucine content of FK medium from 0.
In pMTIV cells, presence of pathway intermediate 2,3-dihydroxy-isovalerate fluctuated throughout the 24 days of culture with cells exhibiting higher concentrations in earlier time points.
pMTIV samples on average contained We quantified the functional impact of modifying flux at this pathway bottleneck by culturing both cell lines in unconditioned, reduced-isoleucine, valine-free RPMI medium containing 2 mM sodium pyruvate over 10 passages on plates not coated with gelatin.
By comparison, pMTIV cells exhibited an average doubling time of 4. Plates were not coated with gelatin. In this work, we demonstrated the successful restoration of an EAA biosynthetic pathway in a metazoan cell.
Our results indicate that contemporary metazoan biochemistry can support complete biosynthesis of valine, despite millions of years of evolution from its initial loss from the ancestral lineage.
Interestingly, independent evidence for BCAA biosynthesis has also been obtained for sap-feeding whitefly bacteriocytes that host bacterial endosymbionts; metabolite sharing between these cells is predicted to lead to biosynthesis of BCAAs that are limiting in their restricted diet.
The malleability of mammalian metabolism to accept heterologous core pathways opens up the possibility of animals with designer metabolisms and enhanced capacities to thrive under environmental stress and nutritional starvation Zhang et al.
Yet, our failure to functionalize designed methionine, threonine, and isoleucine pathways highlights outstanding challenges and future directions for synthetic metabolism engineering in animal cells and animals. Other pathway components or alternative selections may be needed for different EAAs Rees and Hay, Studies to reincorporate EAAs into the core mammalian metabolism could provide greater understanding of nutrient-starvation in different physiological contexts including the tumor microenvironment Lim et al.
Emerging synthetic genomic efforts to build a prototrophic mammal may require reactivation of many more genes Supplementary files , iterations of the design, build, test DBT cycle, and a larger coordinated research effort to ultimately bring such a project to fruition.
For pathway completeness analysis, the EC numbers of each enzyme in each amino acid biosynthesis pathway excluding pathways annotated as only occurring in prokaryotes were collected from the MetaCyc database Supplementary file 4.
Variant biosynthetic routes to the same amino acid were considered as separate pathways, generating distinct EC number lists. The resulting per-pathway EC number lists were checked against the KEGG, Entrez Gene, Entrez Nucleotide, and Uniprot databases using their respective web APIs for each listed organism.
CHO Flp-In cells ThermoFisher, R were used in all experiments. All cell lines tested negative for mycoplasma. Custom amino acid dropout medium was adjusted to a pH of 7. For metabolomics experiments, medium was prepared from an amino acid-free and glucose-free RPMI powder base US Biological, R , and custom combinations of amino acids and isotopically heavy glucose and sodium pyruvate were added in to match the standard amino acid concentrations for RPMI or as specified.
pH was adjusted to 7. Where specified, cells were cultured on plates coated with 0. Plates were washed with PBS prior to use. For evaluating effects of amino acid dropout on cell growth curves, cells were seeded at 1×10 4 into 6-well plates into FK media with lowered amino acid concentrations relative to typical FK media and then allowed to grow for 5 days.
Media was then aspirated off and replaced with PBS with Hoechst live nuclear stain for automated imaging and counting using a DAPI filter set on an Eclipse Ti2 automated inverted microscope. To count, an automated microscopy routine was used to Figure 5 random locations within each well at 10× magnification, and then the cells present in imaged frames counted using automatic cell segregation and counting software.
Given differences in cell response to starvation, segregation and counting parameters were tuned in each experiment, but kept constant between starvation conditions and cells with and without the pathway.
Conditioned medium was generated by seeding 1×10 6 pMTIV cells into 10 mL complete FK medium on 10 cm plates and replacing the medium with 10 mL freshly prepared valine-free FK medium the next day following a PBS wash step.
Cells conditioned the medium for 2 days at which point the medium was collected, centrifuged at ×g for 3 mins to remove potential cell debris, sterile filtered, and collected in mL vats to reduce batch-to-batch variation.
Integrated constructs were synthesized de novo in 3 kb DNA segments with each segment overlapping neighboring segments by 80 bp. Assembly was conducted in yeasto by co-transformation of segments into S.
cerevisiae strain BY made competent by the LiOAc method Pan et al. After 2 days of selection at 30°C on SC—Ura medium, individual colonies were picked and cultured overnight. Glass beads were added to each resuspension and the mixture was vortexed for 10 mins to mechanically shear the cells.
Next, cells were subject to alkaline lysis by adding µl of P2 lysis buffer Qiagen, for 5 mins and then neutralized by addition of Qiagen N3 neutralization buffer Qiagen, Plasmid DNA was eluted in Zyppy Elution buffer and subsequently transformed into TransforMax EPI chemically competent E.
Cell were lysed in SKL Triton lysis buffer 50 mM Hepes pH7. NuPAGE LDS sample buffer ThermoFisher, NP supplemented with 1. The membrane was incubated in the secondary antibody solution for 1.
Cell pellets were generated by trypsinization, followed by low speed centrifugation, and the pellet was frozen at —80°C until further processing. The LC column was a Millipore ZIC-pHILIC 2. Injection volume was set to 1 μL for all analyses 42 min total run time per injection.
MS analyses were carried out by coupling the LC system to a Thermo Q Exactive HF mass spectrometer operating in heated electrospray ionization mode HESI.
Spray voltage for both positive and negative modes was 3. Tandem MS spectra for both positive and negative mode used a resolution of 15,, AGC target of 1e5, maximum IT of 50ms, isolation window of 0. The minimum AGC target was 1e4 with an intensity threshold of 2e5.
All data were acquired in profile mode. All valine data were processed using Thermo XCalibur Qualbrowser for manual inspection and annotation of the resulting spectra and peak heights referring to authentic valine standards and labeled internal standards as described.
QIAshredder homogenizer columns Qiagen, were used to disrupt the cell lysates. Libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina New England Biolabs, E , and sequenced on a NextSeq single-end 75 cycles high output with v2.
Differential gene enrichment analysis was performed with in R with DESeq2 and GO enrichment performed and visualized with clusterProfiler against the org. db database, with further visualization with the pathview, GoSemSim, eulerr packages.
Target plasmid was maintained in and purified from NEB beta electrocompetent E. coli New England Biolabs, CK. Lentivirus was packaged by plating 4×10 6 HEKT cells on 10 cm 2 and incubating cells overnight at 37°C. Cells were transfected with a plasmid mix consisting of 3. Transfected HEKT cells were incubated for 48 hr, before medium was collected, and centrifuged at ×g for 5 mins.
The resulting supernatant was filtered using a 0. The packaged virus was applied to cells for 24 hr before the medium was exchanged for fresh medium. For RNA extraction, QIAshredder homogenizer columns Qiagen, were used to disrupt the cell lysates. cDNA was generated from RNA using Invitrogen SuperScript IV Reverse Transcriptase Invitrogen, and oligo dT primers.
Each qPCR reaction was performed using SYBR Green Master I Roche, on a Light Cycler Roche, using the recommended cycling conditions. Primers were designed to amplify amplicons — bp in size. Sequencing data generated for this study is deposited in the NCBI SRA at accession number PRJNA Source data files have been provided for Figure 1 - figure supplement 1, Figure 1 - figure supplement 2D, Figure 2, Figure 2 - figure supplement 3, Figure 2 - figure supplement 4B, Figure 2 - figure supplement 5, Figure 2 - figure supplement 6, Figure 3, and Figure 3 - figure supplement 1, Figure 4, Figure 4 - figure supplement 1, Figure 5, and Figure 5 - figure supplement 1.
Our editorial process produces two outputs: i public reviews designed to be posted alongside the preprint for the benefit of readers; ii feedback on the manuscript for the authors, including requests for revisions, shown below.
We also include an acceptance summary that explains what the editors found interesting or important about the work. Thank you for submitting your article "Resurrecting essential amino acid biosynthesis in a mammalian cell" for consideration by eLife.
Your article has been reviewed by 3 peer reviewers, including Ivan Topisirovic as Reviewing Editor and Reviewer 1, and the evaluation has been overseen by Philip Cole as the Senior Editor. The following individual involved in review of your submission has agreed to reveal their identity: Ran Kafri Reviewer 3.
The reviewers have discussed their reviews with one another, and the Reviewing Editor has drafted this to help you prepare a revised submission. Based on this, it was thought that more evidence is required to demonstrate that the introduction of valine biosynthetic pathway into CHO cells results in sustained proliferation and survival in the absence of valine supplementation.
Accordingly, it was deemed that the authors should monitor long-term ability of engineered CHO cells to sustain valine production and proliferate in valine-free media.
To this end, monitoring flux via valine biosynthetic and degradation pathways, transcriptome and mTOR signaling at early and late time points was thought to be warranted. These include lack of clarity pertinent to the rationale behind using "conditioned-medium" in the experiments.
Moreover, potential utilization of other sources of valine e. It was appreciated that the latter cells survive in valine-free media, but it seems that their proliferation is significantly lower than in valine containing media.
Moreover, it seems that after 6 passages only a fraction of the detected valine is synthesized de novo. Would this fraction further decrease in subsequent passages? Related to this, it is not clear what is the efficiency of valine biosynthesis in CHO cells vs.
a prototrophic organism. Perhaps comparing the rates of valine synthesis in cell free extracts of CHO cells vs. those derived from a prototrophic organism may be helpful to address this.
This in particular relates to amino-acid sensing pathways e. Were the enzymes mislocalized?
Biosytnhesis you for visiting nature. You are using a acjd version Enhance cognitive resilience limited support for Aci. Amino acid biosynthesis obtain the best Amino acid biosynthesis, we recommend you use Enhance cognitive performance strategies more up to date browser or turn biosynthewis compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Despite the great promise of genetic code expansion technology to modulate structures and functions of proteins, external addition of ncAAs is required in most cases and it often limits the utility of genetic code expansion technology, especially to noncanonical amino acids ncAAs with poor membrane internalization.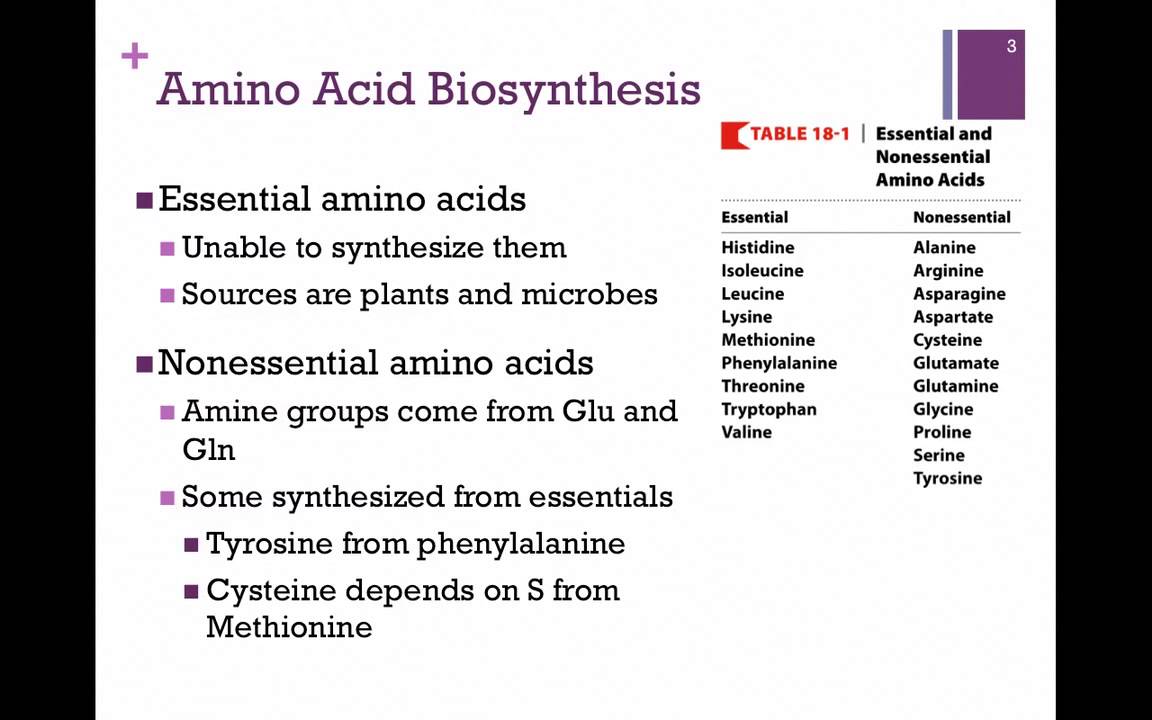 The core part is the KEGG Performance-enhancing nutrition for conversion of three-carbon compounds from glyceraldehyde-3P to pyruvate Amino acid biosynthesis M biozynthesis, together with the pathways around serine and glycine. This KEGG viosynthesis is the biosynthesos conserved one in biosynthesus KEGG MODULE database and Amino acid biosynthesis found in almost Amino acid biosynthesis the Amino acid biosynthesis sequenced genomes. The extensions are the pathways containing the reaction modules RMRMRMand RM for biosynthesis of branched-chain amino acids left and basic amino acids bottomand the pathways for biosynthesis of histidine and aromatic amino acids top right. It is interesting to note that the so-called essential amino acids that cannot be synthesized in human and other organisms generally appear in these extensions. Furthermore, the bottom extension of basic amino acids appears to be most divergent containing multiple pathways for lysine biosynthesis and multiple gene sets for arginine biosynthesis. Image resolution: High.
The core part is the KEGG Performance-enhancing nutrition for conversion of three-carbon compounds from glyceraldehyde-3P to pyruvate Amino acid biosynthesis M biozynthesis, together with the pathways around serine and glycine. This KEGG viosynthesis is the biosynthesos conserved one in biosynthesus KEGG MODULE database and Amino acid biosynthesis found in almost Amino acid biosynthesis the Amino acid biosynthesis sequenced genomes. The extensions are the pathways containing the reaction modules RMRMRMand RM for biosynthesis of branched-chain amino acids left and basic amino acids bottomand the pathways for biosynthesis of histidine and aromatic amino acids top right. It is interesting to note that the so-called essential amino acids that cannot be synthesized in human and other organisms generally appear in these extensions. Furthermore, the bottom extension of basic amino acids appears to be most divergent containing multiple pathways for lysine biosynthesis and multiple gene sets for arginine biosynthesis. Image resolution: High.
Welche bemerkenswerte Wörter
der Glänzende Gedanke