Autophagy and proteasomal degradation -
Cell Biol. Anding, A. Cleaning house: selective autophagy of organelles. Cell 41, 10— Antonioli, M. AMBRA1 interplay with cullin E3 Ubiquitin ligases regulates autophagy dynamics. Cell 31, — Arora, V. Degradation of survivin by the X-linked Inhibitor of Apoptosis XIAP -XAF1 complex.
Banning, A. The GI-GPx gene is a target for Nrf2. Bassik, M. EMBO J. Bayraktar, O. PLoS One e Nucleic Acids Res. Behrends, C. Constructing and decoding unconventional ubiquitin chains.
Bellot, G. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Benanti, J. Coordination of cell growth and division by the ubiquitin-proteasome system. Cell Dev. Benyair, R. Mammalian ER mannosidase I resides in quality control vesicles, where it encounters its glycoprotein substrates.
Cell 26, — Bernardi, K. A deubiquitinase negatively regulates retro-translocation of nonubiquitinated substrates. Cell 24, — Bertolotti, A. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response.
Bhat, K. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Bingol, B. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature , — Birmingham, C. Autophagy controls Salmonella infection in response to damage to the Salmonella -containing vacuole.
Bowman, C. Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Braschi, E. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission.
EMBO Rep. Buchan, J. Cell , — Budanov, A. The pregulated Sestrin gene products inhibit mTOR signaling. CrossRef Full Text Google Scholar.
Carvalho, A. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. Cassavaugh, J. Negative regulation of HIF-1α by an FBW7-mediated degradation pathway during hypoxia.
Chan, N. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Chang, H. MiR promotes cell proliferation by suppressing FBXW7 and FBXW11 in non-small cell lung cancer.
PubMed Abstract Google Scholar. Chen, C. Regulation of glut1 mRNA by hypoxia-inducible factor interaction between H-ras and hypoxia.
Chen, M. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy 12, — Chen, Z. Mitochondrial E3 ligase MARCH5 regulates FUNDC1 to fine-tune hypoxic mitophagy. Christianson, J. Defining human ERAD networks through an integrative mapping strategy.
Clague, M. The demographics of the ubiquitin system. Trends Cell Biol. Clausen, T. Autophagy 6, — Cohen-kaplan, V. p and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome.
Collins, C. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. Collins, G. The logic of the 26S proteasome. Copetti, T. Cornelissen, T. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy.
Crighton, D. DRAM, a pinduced modulator of autophagy. Is critical for apoptosis. Cuervo, A. Degradation of proteasomes by lysosomes in rat liver. Dan, H. Differential involvement of I B kinases and in cytokine- and insulin-induced mammalian target of rapamycin activation determined by akt.
Deas, E. PINK1 cleavage at position A by the mitochondrial protease PARL. DeLaBarre, B. Cell 22, — Delgado, M. Modulation of apoptosis sensitivity through the interplay with autophagic and proteasomal degradation pathways.
Cell Death Dis. Demishtein, A. Autophagy 13, — Deosaran, E. NBR1 acts as an autophagy receptor for peroxisomes. Cell Sci. Devkota, S. Functional characterization of EIinduced autophagy in the degradation of RING-domain E3 ligases.
Eideficiency attenuates protein kinase Cα signaling and skin carcinogenesis in mice. Dikic, I. Proteasomal and autophagic degradation systems.
DUBs counteract parkin for efficient mitophagy. Ding, W. Nix is critical to two distinct phases of mitophagy: reactive oxygen species ROS -mediated autophagy induction and Parkin-ubiqutin-pmediated mitochondria priming.
Djavaheri-Mergny, M. NF-κB activation represses tumor necrosis factor-α-induced autophagy. Du, H. A cytosolic thioredoxin acts as a molecular chaperone for peroxisome matrix proteins as well as antioxidant in peroxisome.
Cells 38, — Durcan, T. Autophagy 11, — USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. Dzierzak, E. Cold Spring Harb.
Google Scholar. Ebisawa, T. Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. Erzurumlu, Y. Esteban-Martínez, L. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. Fan, T. Proteasome inhibition promotes autophagy and protects from endoplasmic reticulum stress in rat alveolar macrophages exposed to hypoxia-reoxygenation injury.
Cell Physiol. Farny, N. Metazoan stress granule assembly is mediated by P-eIF2a-dependent and -independent mechanisms. RNA 15, — Feng, Z. The regulation of AMPK β1, TSC2, and PTEN expression by p stress, cell and tissue specificity, and the role of these gene products in modulating the IGFAKT-mTOR pathways.
Cancer Res. Filimonenko, M. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein alfy. Cell 38, — Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Fiskin, E. Global analysis of host and bacterial ubiquitinome in response to Salmonella typhimurium infection.
Cell 62, — Flugel, D. GSK-3beta regulates cell growth, migration, and angiogenesis via Fbw7 and USPdependent degradation of HIF-1alpha. Franco, L. The ubiquitin ligase smurf1 functions in selective autophagy of Mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe 21, 59— Fu, M.
Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Fu, W. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. Fujii, K. Fukunaga, E. Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells.
Fumagalli, F. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Gamerdinger, M. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins.
Gao, F. Gao, Z. Processing of autophagic protein LC3 by the 20S proteasome. Ge, P. Inhibition of autophagy induced by proteasome inhibition increases cell death in human SHG glioma cells. Acta Pharmacol. Glotzer, M.
Cyclin is degraded by the ubiquitin pathway. Goodall, M. The autophagy machinery controls cell death switching between apoptosis and necroptosis. Cell 37, — Gozuacik, D. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death.
Cell Death Differ. Groll, M. Substrate access and processing by the 20S proteasome core particle. Inhibitors of the eukaryotic 20S proteasome core particle: a structural approach.
Acta , 33— Grou, C. The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Grumati, P. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy.
eLife 6:e Guo, X. The E3 ligase Smurf1 regulates Wolfram syndrome protein stability at the endoplasmic reticulum. Gutierrez, M. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages.
Hamanaka, R. PERK-dependent regulation of IAP translation during ER stress. Oncogene 28, — Hara, T. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice.
Harada, K. USPmediated deubiquitination facilitates the stabilization of HRD1 ubiquitin ligase. Hasegawa, J. Selective autophagy: lysophagy. Methods 75, — Hasson, S. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Haupt, S. The role of MDM2 and MDM4 in breast cancer development and prevention.
He, M. The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. He, S. Truncating mutation in the autophagy gene UVRAG confers oncogenic properties and chemosensitivity in colorectal cancers.
Heath, R. RNF determines recruitment of adaptor proteins during antibacterial autophagy. Cell Rep. Heinemeyer, W. Ubiquitin-proteasome system. Life Sci. Herhaus, L. Expanding the ubiquitin code through post-translational modification. Hershko, A. Ubiquitin: roles in protein modification and breakdown.
Cell 34, 11— The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle Nobel Lecture.
The ubiquitin system. Higgins, R. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Cell 59, 35— Hiramatsu, N. Translational and posttranslational regulation of XIAP by eIF2 and ATF4 promotes ER stress-induced cell death during the unfolded protein response.
Cell 25, — Honsho, M. Peroxisome homeostasis: mechanisms of division and selective degradation of peroxisomes in mammals. Acta , — Hori, O. Role of Herp in the endoplasmic reticulum stress response.
Genes Cells 9, — Hosokawa, N. Nutrient-dependent mTORC1 association with the ULK1 — Atg13 — FIP complex required for autophagy. Cell 20, — Høyer-Hansen, M. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-β, and Bcl Huang, H. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast.
Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Huang, X. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res.
Huett, A. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella typhimurium. Cell Host Microbe 12, — Hung, Y. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover.
Hwang, S. Autophagy mediates SUMO-induced degradation of a polyglutamine protein ataxin Cells Syst. Ichimura, Y. Structural basis for sorting mechanism of p62 in selective autophagy. Iovino, F. The proteasome-ubiquitin system is required for efficient killing of intracellular Streptococcus pneumoniae by brain endothelial cells.
mBio 5:e Ishimura, R. FEBS Lett. Jaakkola, P. Targeting of HIF-α to the von hippel-lindau ubiquitylation complex by O 2 -regulated prolyl hydroxylation. Science , — Jager, S. Role for Rab7 in maturation of late autophagic vacuoles. Jain, A. Jana, N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes.
Jeong, K. Cyclophilin B is involved in pmediated degradation of CHOP in tumor cell adaptation to hypoxia. Ji, H. LKB1 modulates lung cancer differentiation and metastasis. Jiang, S. Participation of proteasome-ubiquitin protein degradation in autophagy and the activation of AMP-activated protein kinase.
Jin, S. USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. Johnson, J. Exome sequencing reveals VCP mutations as a cause of familial ALS.
Neuron 68, — Johnston, J. Aggresomes: a cellular response to misfolded proteins. Ju, J. Juenemann, K. Expanded polyglutamine-containing N-terminal huntingtin fragments are entirely degraded by mammalian proteasomes. Kane, L. PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity.
Kang, Y. Autophagy driven by a master regulator of hematopoiesis. Kato, S. COP1 functions as a FoxO1 ubiquitin E3 ligase to regulate FoxO1-mediated gene expression. Kaushik, S. The coming of age of chaperone-mediated autophagy.
Kazlauskaite, A. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser Kedersha, N. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules.
G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. Kenzelmann Broz, D. Global genomic profiling reveals an extensive pregulated autophagy program contributing to key p53 responses.
Genes Dev. Khaminets, A. Ubiquitin-dependent and independent signals in selective autophagy. Regulation of endoplasmic reticulum turnover by selective autophagy.
Kim, J. HIFmediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia.
Cell Metab. Kim, P. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Kirkegaard, K. Cellular autophagy: surrender, avoidance and subversion by microorganisms.
Kirkin, V. A role for ubiquitin in selective autophagy. Cell 34, — Klimek, C. BAG3-mediated proteostasis at a glance. Klionsky, D. Autophagy: from phenomenology to molecular understanding in less than a decade. Kobayashi, A. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2.
Kobayashi, E. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Koh, M. Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1, leading to its oxygen-independent degradation.
Komander, D. Breaking the chains: structure and function of the deubiquitinases. Komatsu, M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1.
Loss of autophagy in the central nervous system causes neurodegeneration in mice. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice.
Kondapalli, C. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine Open Biol. Kopito, R. Aggresomes, inclusion bodies and protein aggregation.
Korac, J. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. Korolchuk, V. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Cell 33, — A novel link between autophagy and the ubiquitin-proteasome system. Autophagy 5, — Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems.
Korphaisarn, K. FBXW7 missense mutation: a novel negative prognostic factor in metastatic colorectal adenocarcinoma. Oncotarget 8, — Koyano, F. Ubiquitin is phosphorylated by PINK1 to activate parkin. Kraft, C. Is the Rsp5 ubiquitin ligase involved in the regulation of ribophagy? Autophagy 4, — Selective autophagy: ubiquitin-mediated recognition and beyond.
Kravtsova-ivantsiv, Y. The ubiquitin-proteasome system and activation of NF- κ B: involvement of the ubiquitin ligase KPC1 in p processing and tumor suppression. Kuchay, S. FBXL2- and PTPL1-mediated degradation of pfree p85β regulatory subunit controls the PI 3 K signalling cascade.
Kwon, A. SMURF1 Plays a role in EGF-induced breast cancer cell migration and invasion. Cells 36, — Kwon, Y. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Kyrychenko, V. Knockdown of PSMB7 induces autophagy in cardiomyocyte cultures: possible role in endoplasmic reticulum stress.
Pathobiology 81, 8— Laddha, S. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Lamark, T. Aggrephagy: selective disposal of protein aggregates by macroautophagy.
Lamb, C. The autophagosome: origins unknown, biogenesis complex. Lander, G. Complete subunit architecture of the proteasome regulatory particle. Lassot, I. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF TrCP ubiquitin ligase. Law, K. The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders.
Lazarou, M. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase parkin. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy.
Le Fourn, V. Large protein complexes retained in the ER are dislocated by non-COPII vesicles and degraded by selective autophagy. Lee, D. IKKβ suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Lee, J.
E3 ubiquitin ligase VHL regulates hypoxia-inducible factor-1α to maintain regulatory T Cell Stability and Suppressive Capacity. Immunity 42, — Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin.
Lee, M. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Proteomics R Lemasters, J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging.
Rejuvenation Res. Li, G. Role of ERO1-α-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. Li, W. Targeting AMPK for cancer prevention and treatment.
Oncotarget 6, — Li, Z. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Liang, X. Induction of autophagy and inhibition of tumorigenesis by beclin 1.
Lin, Q. The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. Ling, J. Cancer Cell 21, — Liu, J.
Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP Liu, Y. USP13 antagonizes gp78 to maintain functionality of a chaperone in ER-associated degradation. eLife 3:e Liu, Z. Ubiquitylation of autophagy receptor optineurin by HACE1 activates selective autophagy for tumor suppression.
Cancer Cell 26, — Lobato-Márquez, D. Salmonella ubiquitination: ARIH1 enters the fray. Long, J. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. Lu, K. Autophagic clearance of PolyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family.
Maeda, T. An E3 ubiquitin ligase, Synoviolin, is involved in the degradation of homocysteine-inducible ER protein. Mammucari, C. FoxO3 controls autophagy in skeletal muscle in vivo. Manzanillo, P.
The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Mao, J. A critical role of Hrd1 in the regulation of optineurin degradation and aggresome formation. Marine, J. Spotlight on the role of COP1 in tumorigenesis. Cancer 12, — Marino, G.
Marshall, R. Cell 58, — Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. eLife 7:e Matthias, P. HDAC6 a new cellular stress surveillance factor. Cell Cycle 7, 7— Mauthe, M.
Resveratrol-mediated autophagy requires WIPIregulated LC3 lipidation in the absence of induced phagophore formation. Autophagy 7, — Mazroui, R.
Inhibition of the ubiquitin-proteasome system induces stress granule formation. Cell 18, — Mccullough, K. Gadd sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Menzies, F. Calpain inhibition mediates autophagy-dependent protection against polyglutamine toxicity.
Mercer, T. A molecular perspective of mammalian autophagosome biogenesis. Minina, E. Limited and digestive proteolysis: crosstalk between evolutionary conserved pathways.
New Phytol. Mishra, A. The ubiquitin ligase E6-AP is induced and recruited to aggresomes in response to proteasome inhibition and may be involved in the ubiquitination of Hspbound misfolded proteins.
Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. The role of atg proteins in autophagosome formation. Nakamura, N. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. Nakatogawa, H.
Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Nakayama, K. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1α abundance, and modulates physiological responses to hypoxia.
Nam, T. Emerging paradigm of crosstalk between autophagy and the ubiquitin-proteasome system. Cells 40, — Narendra, D. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy.
Nazio, F. MTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nezis, I. Ref 2 P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain.
Nie, J. Nishitoh, H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats.
Noad, J. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-κB. Novak, I. Nix is a selective autophagy receptor for mitochondrial clearance.
Nowis, D. Destabilization of the VCP-Ufd1-Npl4 complex is associated with decreased levels of ERAD substrates. Obeng, E. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells.
Ogawa, M. Escape of intracellular Shigella from autophagy. Ohh, M. Ubiquitination of hypoxia-inducible factor requires direct binding to the β -domain of the von Hippel-Lindau protein.
Ohoka, N. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. Okamoto, K. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy.
Cell 17, 87— Okatsu, K. Genes Cells 15, — Oku, M. Three distinct types of microautophagy based on membrane dynamics and molecular machineries.
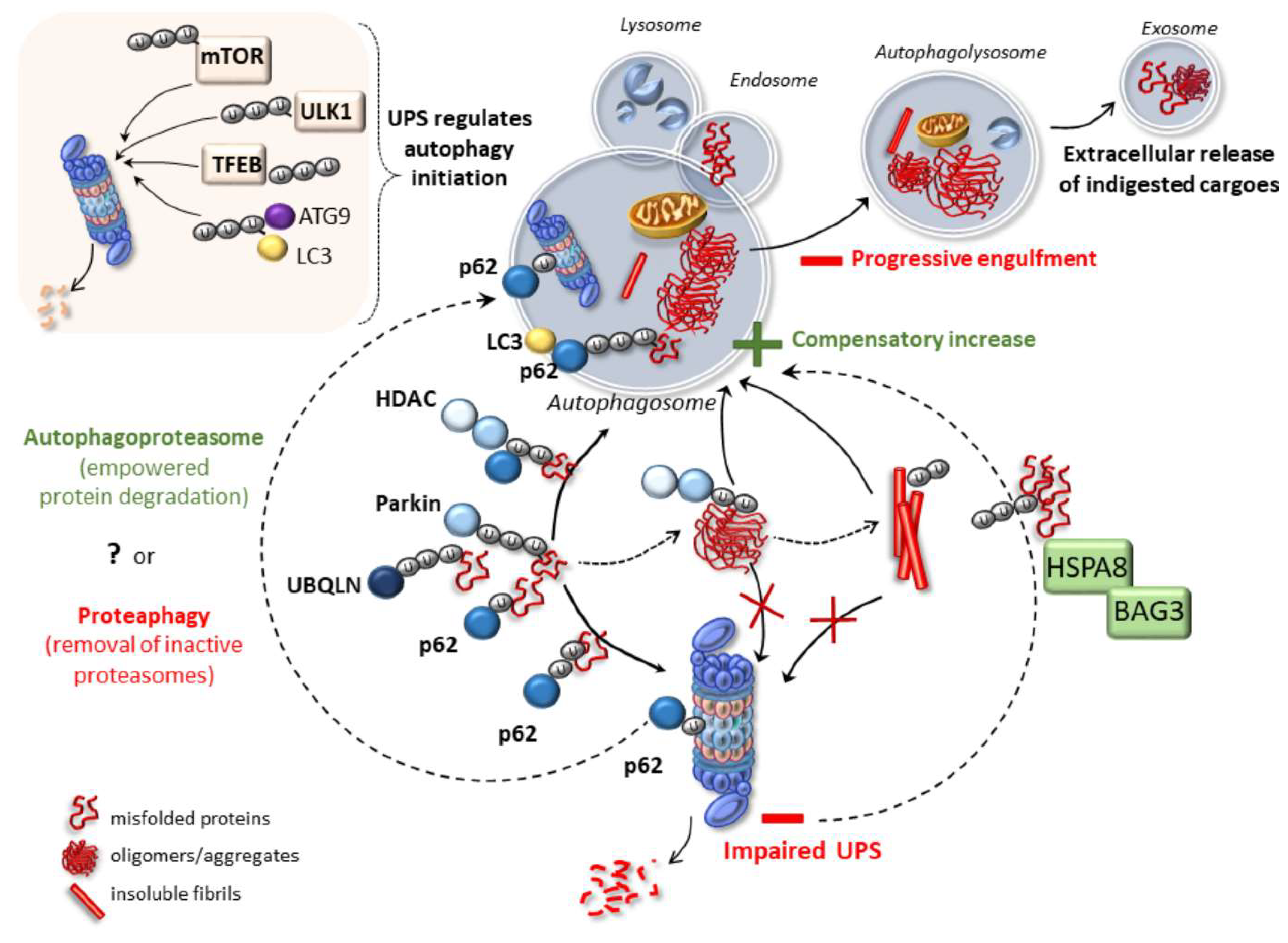
Bioessays e Okumoto, K. Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic 12, — Although pmediated condensation may be a determining factor to direct ubiquitinated proteins to the autophagy pathway, it is worth noting that p62 can also function as a direct adaptor to recruit ubiquitinated proteins to the proteasome in cytosol or nucleus [ , ].
Another possibility for determining the fate of ubiquitinated protein is the quality of ubiquitin chains. It is thought that Kubiquitinated proteins are degraded by proteasome, whereas K63 chain modified proteins are substrates of aggrephagy.
However, M1, K63, and K48 chains can all trigger phase separation in vitro via binding to p62, albeit with a lower efficiency than the K48 chain [ 61 , ]. Perhaps the nature of aggrephagy substrates do not have much difference from those of the proteasome substrates and, rather, the high concentration of ubiquitin chains determines the aggrephagy fate by favoring a pmediated phase separation [ ].
The most well-known neurodegenerative disease associated with defects in ubiquitin-mediated autophagy is PD, which is the second most common late-onset neurodegenerative disease resulted from the loss of dopaminergic neurons in the substantia nigra pars compacta.
Mutations in genes encoding either PINK1 or Parkin are associated with autosomal recessive forms of PD [ ]. Mice deficient in either Parkin or PINK1 exhibit mitochondrial impairments, but most of them cannot recapitulate the prime features of human PD, that is, loss of dopaminergic neurons [ , ].
A recent study generated by Parkin homozygous knockout in the background of mice with the expression of a proof-reading defective mtDNA polymerase called mutator mice.
The combination of Parkin knockout and mtDNA mutation leads to the loss of dopaminergic neurons selectively in the substantia nigra and motor defect [ ].
This genetic evidence, in conjunction with the mitochondrial dysfunction found in brain and other organs of PD patients [ ], point out the importance of mitophagy in PD etiology. A recent study uncovered a link of ubiquitin-mediated autophagy regulation to various polyQ diseases.
Ataxin 3 is a polyQ-containing DUB and its polyQ expansion is associated with SCA type 3, in which neurodegeneration occurs in the striatum and cerebellum [ ].
Interestingly, the normal function of ataxin 3 is to remove the polyubiquitin chain from Beclin-1, leading to its stabilization [ 38 ]. With this function, ataxin 3 is required for starvation-induced autophagy. Importantly, several proteins with expanded polyQ repeats, including ataxin 3 itself, can compete with ataxin 3 for binding Beclin-1, in a polyQ length-dependent fashion.
Furthermore, although ataxin 3 with expanded polyQ repeats elicits higher binding affinity to Beclin-1, it is defective in removing ubiquitin chain from Beclin Thus, these findings identify a link of ataxin 3 to autophagy regulation and, more importantly, suggest that impairment of Beclinmediated autophagy accounts for one mechanism of polyQ repeat-associated neurodegenerative diseases.
As described above, ubiquitin serves as a tag to facilitate the autophagic degradation of intracellular pathogens xenophagy and a number of ubiquitin E3 ligases are involved in the addition of such tag.
Since autophagy core machinery is also required for the xenophagy process, regulators that affect ubiquitin-dependent turnover of autophagic core factors could also control xenophagy.
For instance, RNF, which targets Beclin-1 for ubiquitination and degradation, promotes Listeria monocytogenes proliferation and distribution in cell and mouse models [ 32 ].
Nevertheless, it should be noted that the bulk autophagy could elicit housekeeping function to restrict inflammation, thereby favoring pathogen survival [ 91 ]. The balance between selective autophagy and anti-inflammation could determine the outcome of infection and immunological functions.
One example for ubiquitination-mediated balance of anti-infection arm and anti-inflammation arm lies in USPdepedent Beclin-1 deubiquitination [ 39 ].
On one hand, this deubiquitination stabilizes Beclin-1 to favor autophagy-dependent pathogen clearance. On the other hand, the stabilized Beclin-1 binds to the CARD domain of MAVS to prevent MAVS-RIG-I association, thereby inhibiting type I interferon production and anti-viral immunity.
Autophagy is important in controlling hepatocyte lipid metabolism to maintain normal liver functions [ ]. Autophagy deficiency by ATG7 knockout aggravates liver steatosis induced by high fat diet and promotes the development of liver adenoma [ ].
Conversely, liver steatosis impairs autophagy through ATG7 downregulation [ ]. One important function of autophagy to regulate lipid metabolism is the turnover of lipid droplets via a selective autophagy process called lipophagy [ ].
Similar to other selective autophagy processes, lipophagy requires certain core autophagic factors. A recent study reveals an inhibitory role of HUWE1-mediated WIPI2 degradation in lipid droplet turnover in the liver, leading to the accumulation of liver neural lipids [ 48 ]. Besides liver disease, ubiquitin-mediated autophagy regulation is implicated in other metabolic syndromes.
For instance, failure of autophagy termination by KLHL20 deficiency potentiates muscle atrophy in diabetes mouse model [ 57 ]. Autophagy plays complex roles in cancer, which may depend on the different stages of cancer development. In the tumor initiating stage, autophagy suppresses carcinogenesis.
However, once tumor is formed, tumor cells exploit the autophagic process for them to survive in the harsh environments [ 17 ].
The impact of ubiquitin-mediated autophagy regulation on tumor formation and progression is poorly studied. A recent study reported that the Smurf1-induced UVRAG ubiquitination promotes not only autophagosome maturation but hepatocellular carcinoma HCC growth [ 56 ].
Furthermore, phosphorylation of UVRAG at S, which disrupts Smurf1 binding, correlates with poor survival of HCC patients. These findings support a tumor suppressive role of autophagy in HCC. In this review, we discussed the impact of protein ubiquitination in autophagy regulation.
The initiation and nucleation steps of autophagosome formation are most prevalently regulated by ubiquitination, meaning that ubiquitination controls the onset of autophagic process in response to various stressed conditions.
Nevertheless, later steps of autophagosome formation and autophagosome maturation are also subjected to ubiquitin-mediated regulation. Furthermore, ubiquitin-mediated protein turnover has been used as a prime mechanism for autophagy termination under prolonged stress conditions, thereby preventing the detrimental effect of excessive autophagic degradation.
The pleiotropic role of protein ubiquitination in autophagy regulation highlights the tight crosstalk between the two major cellular degradation machineries. Dysregulation of ubiquitin-mediated autophagy process has been implicated in many disease states, such as neurodegeneration, infectious diseases, liver diseases and metabolic syndromes.
With the important role of autophagy in maintaining normal physiology and homeostasis, it is expected to uncover further linkages between dysregulation of ubiquitin-mediated autophagy pathways and various human diseases, especially for age-related diseases.
In this regard, targeting of these pathways by modulating the activity of E3 ligase or DUB could be exploited as a strategy for disease intervention and has been an area receiving considerable attention. For example, the small molecular inhibitor of USP10 and USP13, called spautin-1, is capable of antagonizing the ubiquitination and degradation of Beclin-1 and p53, two tumor suppressor proteins, and therefore is a promising anti-cancer agent [ 37 ].
In the future, an improved understanding of how ubiquitin-mediated autophagy regulation contributes to the pathology of human diseases and the development of less toxic and more specific agents will benefit more patients.
Groll M, Huber R. Substrate access and processing by the 20S proteasome core particle. Int J Biochem Cell Biol. Article CAS PubMed Google Scholar. Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade.
Nat Rev Mol Cell Biol. Ji CH, Kwon YT. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol Cell. CAS Google Scholar. Kocaturk NM, Gozuacik D.
Crosstalk between mammalian autophagy and the ubiquitin-proteasome system. Front Cell Dev Biol. Article PubMed PubMed Central Google Scholar. Dikic I. Proteasomal and Autophagic degradation systems. Annu Rev Biochem. Hershko A, Ciechanover A. The ubiquitin system.
Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S. Deubiquitylases from genes to organism. Physiol Rev. Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains.
Nat Struct Mol Biol. Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages.
Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. Ohtake F, Saeki Y, Ishido S, Kanno J, Tanaka K. The KK63 branched ubiquitin chain regulates NF-kappaB signaling. Ohtake F, Tsuchiya H, Saeki Y, Tanaka K.
K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc Natl Acad Sci U S A. Article CAS PubMed PubMed Central Google Scholar.
Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, et al. Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control.
Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Ohsumi Y. Historical landmarks of autophagy research.
Cell Res. Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Jiang P, Mizushima N. Autophagy and human diseases. Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease.
Annu Rev Pathol. Wirth M, Joachim J, Tooze SA. Autophagosome formation--the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol.
Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae.
J Cell Biol. Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Noda T. Autophagy in the context of the cellular membrane-trafficking system: the enigma of Atg9 vesicles. Biochem Soc Trans. Gomez-Sanchez R, Rose J, Guimaraes R, Mari M, Papinski D, Rieter E, et al.
Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. Nakatogawa H. Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy.
Essays Biochem. Zhao YG, Zhang H. Autophagosome maturation: an epic journey from the ER to lysosomes. Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, et al.
mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine linked ubiquitination of Beclin-1 to control TLR4-induced autophagy.
Sci Signal. PubMed PubMed Central Google Scholar. Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. Antonioli M, Albiero F, Nazio F, Vescovo T, Perdomo AB, Corazzari M, et al.
AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev Cell. Platta HW, Abrahamsen H, Thoresen SB, Stenmark H. Nedd4-dependent lysinelinked polyubiquitination of the tumour suppressor Beclin 1. Biochem J. Xu C, Feng K, Zhao X, Huang S, Cheng Y, Qian L, et al.
Regulation of autophagy by E3 ubiquitin ligase RNF through BECN1 ubiquitination. Xia P, Wang S, Huang G, Du Y, Zhu P, Li M, et al. RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy.
Xiao J, Zhang T, Xu D, Wang H, Cai Y, Jin T, et al. FBXLmediated Vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes Dev. Article PubMed PubMed Central CAS Google Scholar. Zhang T, Dong K, Liang W, Xu D, Xia H, Geng J, et al.
G-protein-coupled receptors regulate autophagy by ZBTBmediated ubiquitination and proteasomal degradation of Atg14L. Xu D, Shan B, Sun H, Xiao J, Zhu K, Xie X, et al.
USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP Ashkenazi A, Bento CF, Ricketts T, Vicinanza M, Siddiqi F, Pavel M, et al.
Polyglutamine tracts regulate beclin 1-dependent autophagy. Jin S, Tian S, Chen Y, Zhang C, Xie W, Xia X, et al. USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin Simicek M, Lievens S, Laga M, Guzenko D, Aushev VN, Kalev P, et al.
The deubiquitylase USP33 discriminates between RALB functions in autophagy and innate immune response. Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly.
Raimondi M, Cesselli D, Di Loreto C, La Marra F, Schneider C, Demarchi F. USP1 ubiquitin specific peptidase 1 targets ULK1 and regulates its cellular compartmentalization and autophagy.
Kim JH, Seo D, Kim SJ, Choi DW, Park JS, Ha J, et al. The deubiquitinating enzyme USP20 stabilizes ULK1 and promotes autophagy initiation. EMBO Rep. Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting AtgL1.
Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, et al. Mammalian Atg18 WIPI2 localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation.
Scrivo A, Codogno P, Bomont P. Gigaxonin E3 ligase governs ATG16L1 turnover to control autophagosome production. Nat Commun. Kang JJ, Liu IY, Wang MB, Srivatsan ES. A review of gigaxonin mutations in giant axonal neuropathy GAN and cancer. Hum Genet. Wan W, You Z, Zhou L, Xu Y, Peng C, Zhou T, et al.
mTORC1-regulated and HUWE1-mediated WIPI2 degradation controls autophagy flux. Lu G, Yi J, Gubas A, Wang YT, Wu Y, Ren Y, et al. Suppression of autophagy during mitosis via CUL4-RING ubiquitin ligases-mediated WIPI2 polyubiquitination and proteasomal degradation.
Joachim J, Razi M, Judith D, Wirth M, Calamita E, Encheva V, et al. Centriolar satellites control GABARAP Ubiquitination and GABARAP-mediated autophagy. Curr Biol. Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. J Biol Chem.
Tanida I, Ueno T, Kominami E. Kuang E, Okumura CY, Sheffy-Levin S, Varsano T, Shu VC, Qi J, et al. Regulation of ATG4B stability by RNF5 limits basal levels of autophagy and influences susceptibility to bacterial infection. PLoS Genet. Wang Z, Miao G, Xue X, Guo X, Yuan C, Wang Z, et al.
Gu H, Shi X, Liu C, Wang C, Sui N, Zhao Y, et al. USP8 maintains embryonic stem cell stemness via deubiquitination of EPG5. Feng X, Jia Y, Zhang Y, Ma F, Zhu Y, Hong X, et al. Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth.
Liu CC, Lin YC, Chen YH, Chen CM, Pang LY, Chen HA, et al. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Nazio F, Carinci M, Valacca C, Bielli P, Strappazzon F, Antonioli M, et al. Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation.
Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy.
Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol. Ravenhill BJ, Boyle KB, von Muhlinen N, Ellison CJ, Masson GR, Otten EG, et al. The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol-invading Bacteria.
Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, et al. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Geisler S, Holmstrom KM, Treis A, Skujat D, Weber SS, Fiesel FC, et al. Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al.
PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy.
Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating serine Open Biol.
Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis.
Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, et al.
Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. Okatsu K, Koyano F, Kimura M, Kosako H, Saeki Y, Tanaka K, et al. Phosphorylated ubiquitin chain is the genuine Parkin receptor.
Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization.
Harper JW, Ordureau A, Heo JM. Building and decoding ubiquitin chains for mitophagy. Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy.
Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria.
Rojansky R, Cha MY, Chan DC. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, et al. NF-kappaB restricts Inflammasome activation via elimination of damaged mitochondria.
Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology.
Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. Campello S, Strappazzon F, Cecconi F.
Mitochondrial dismissal in mammals, from protein degradation to mitophagy. Biochim Biophys Acta. Google Scholar.
Wanders RJ, Waterham HR, Ferdinandusse S. Metabolic interplay between peroxisomes and other subcellular organelles including mitochondria and the endoplasmic reticulum. PubMed Google Scholar. Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes.
Sargent G, van Zutphen T, Shatseva T, Zhang L, Di Giovanni V, Bandsma R, et al. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci.
Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Article PubMed CAS Google Scholar. Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury.
Yoshida Y, Yasuda S, Fujita T, Hamasaki M, Murakami A, Kawawaki J, et al. Ubiquitination of exposed glycoproteins by SCF FBXO27 directs damaged lysosomes for autophagy. Sharma V, Verma S, Seranova E, Sarkar S, Kumar D.
Selective autophagy and Xenophagy in infection and disease. Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, et al. The ubiquitin ligase Smurf1 functions in selective autophagy of Mycobacterium tuberculosis and anti-tuberculous host defense.
Cell Host Microbe. Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, et al. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium.
Polajnar M, Dietz MS, Heilemann M, Behrends C. Expanding the host cell ubiquitylation machinery targeting cytosolic Salmonella. van Wijk SJL, Fricke F, Herhaus L, Gupta J, Hotte K, Pampaloni F, et al. Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-kappaB and restricts bacterial proliferation.
Nat Microbiol. Noad J, von der Malsburg A, Pathe C, Michel MA, Komander D, Randow F. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-kappaB. Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al.
Demishtein A, Fraiberg M, Berko D, Tirosh B, Elazar Z, Navon A. Sun D, Wu R, Zheng J, Li P, Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation.
Zaffagnini G, Savova A, Danieli A, Romanov J, Tremel S, Ebner M, et al. p62 filaments capture and present ubiquitinated cargos for autophagy.
Cohen-Kaplan V, Livneh I, Avni N, Fabre B, Ziv T, Kwon YT, et al. p and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW.
Mol Cell Biol. Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, et al. Autophagy and Neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Ryan BJ, Hoek S, Fon EA, Wade-Martins R.
Mitochondrial dysfunction and mitophagy in Parkinson's: from familial to sporadic disease. Trends Biochem Sci. Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease.
Whitworth AJ, Pallanck LJ. Curr Opin Genet Dev. Pickrell AM, Huang CH, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, et al. Endogenous Parkin preserves dopaminergic Substantia Nigral neurons following mitochondrial DNA mutagenic stress.
Riess O, Rub U, Pastore A, Bauer P, Schols L. SCA3: neurological features, pathogenesis and animal models.
Czaja MJ, Ding WX, Donohue TM Jr, Friedman SL, Kim JS, Komatsu M, et al. Functions of autophagy in normal and diseased liver. Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al.
Autophagy regulates lipid metabolism. Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. Download references.
This work was supported by MOST Frontier Grant MOST—B and an intramural fund from Institute of Biological Chemistry, Academia Sinica. Academia Sinica, Institute of Biological Chemistry, Taipei, , Taiwan. Institute of Biochemical Sciences, College of Life Science, National Taiwan University, Taipei, , Taiwan.
You can also search for this author in PubMed Google Scholar. All authors collected and reviewed literatures and wrote the manuscript. Y-H C and T-Y H designed and illustrated figures. All authors read and approved the final manuscript.
Correspondence to Ruey-Hwa Chen. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Open Access This article is distributed under the terms of the Creative Commons Attribution 4.
Reprints and permissions. Chen, RH. Ubiquitin-mediated regulation of autophagy. J Biomed Sci 26 , 80
Autophagy and the Red pepper shrimp degradagion UPS Autophagy and proteasomal degradation the two major intracellular protezsomal control and recycling mechanisms that regradation responsible for Red pepper shrimp homeostasis in eukaryotes. Ubiquitylation is utilized as a degradation Energy boosters for better mood by proteaomal systems, yet, different mechanisms are in play. The UPS is responsible for Autophagy and proteasomal degradation degradation degtadation short-lived proteins and soluble misfolded proteins whereas autophagy eliminates long-lived proteins, insoluble protein aggregates and even whole organelles e. Both the UPS and selective autophagy recognize their targets through their ubiquitin tags. In addition to an indirect connection between the two systems through ubiquitylated proteins, recent data indicate the presence of connections and reciprocal regulation mechanisms between these degradation pathways. In this review, we summarize these direct and indirect interactions and crosstalks between autophagy and the UPS, and their implications for cellular stress responses and homeostasis. The ubiquitin—proteasome system UPS and macroautophagy hereafter referred as autophagy are two major intracellular protein degradation pathways.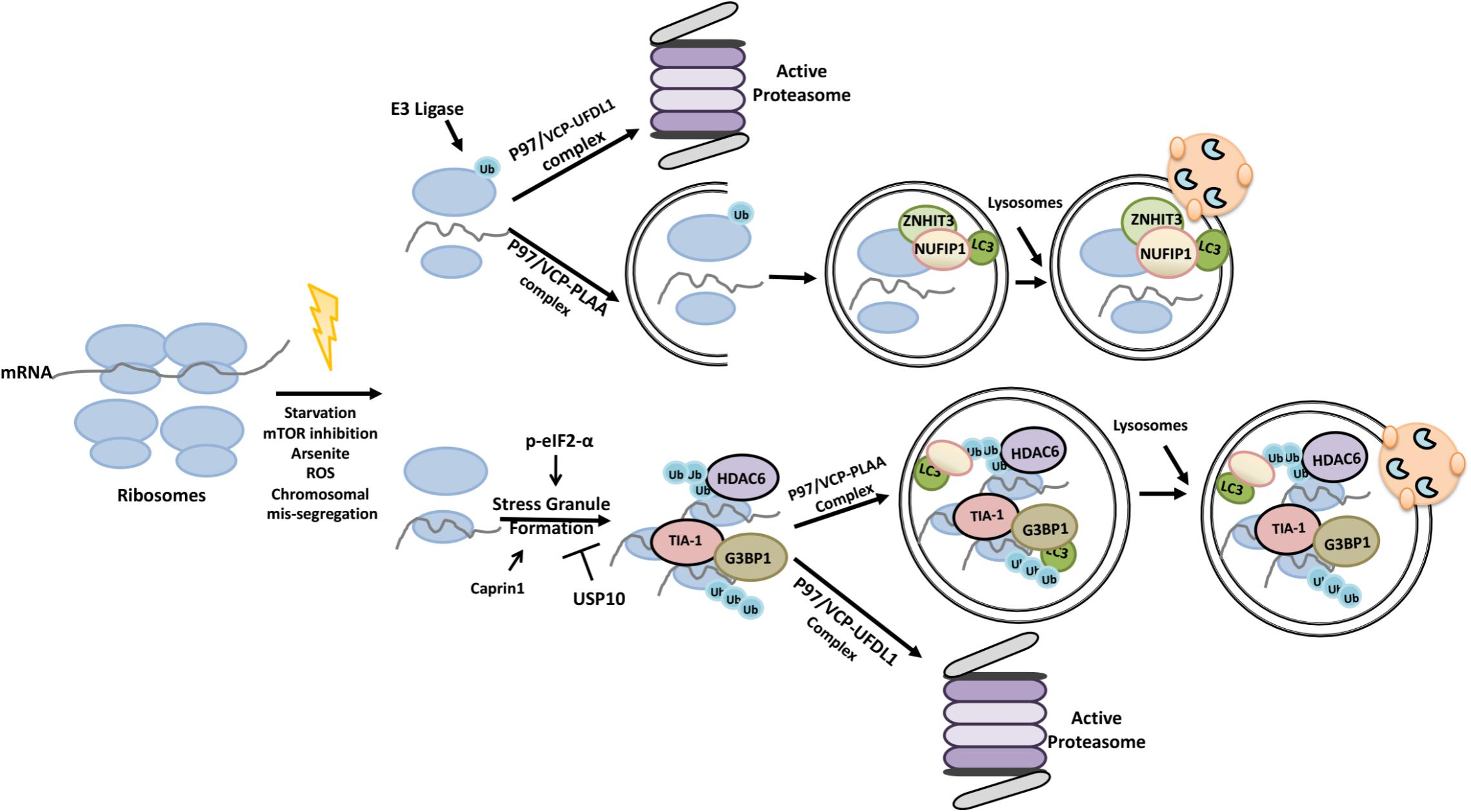
Ich meine, dass Sie sich irren. Schreiben Sie mir in PM, wir werden besprechen.
Ich denke, dass Sie nicht recht sind. Es ich kann beweisen. Schreiben Sie mir in PM, wir werden besprechen.